








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 182-194.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0216
LI Yu-zhen1( ), LI Meng-dan1(
), LI Meng-dan1( ), ZHANG Wei1, PENG Ting2(
), ZHANG Wei1, PENG Ting2( )
)
Received:2025-03-02
Online:2025-09-26
Published:2025-09-24
Contact:
PENG Ting
E-mail:1115662345@qq.com;tpeng@mail.hzau.edu.cn
LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp.[J]. Biotechnology Bulletin, 2025, 41(9): 182-194.
| Gene name | Forward primer (5′‒3′) | Reverse primer (5′‒3′) |
|---|---|---|
| RmUBC | GCCAGAGATTGCCCATATGTA | TCACAGAGTCCTAGCAGCACA |
| AtEF1α | GCAAGATGGATGCCACTACCC | GCAAGATGGATGCCACTACCC |
| RmEXPB2 | TCTAGAATGGCTAGTTTAAGAAGCAT | CCCGGGTAGATAATTGACCACGGATC |
| RmEXPB2-qRT | TGCTGCTTGTTTGAGCAGAG | AAGCTCCACATTCCCTGCC |
| RmEXPB2-eGFP | GAGAACACGGGGGACTCTAGAATGGCTAGTTTAAGAAGCAT | GCCCTTGCTCACCATCCCGGGTAGATAATTGACCACGGATC |
Table 1 Primer sequences for RT-qPCR
| Gene name | Forward primer (5′‒3′) | Reverse primer (5′‒3′) |
|---|---|---|
| RmUBC | GCCAGAGATTGCCCATATGTA | TCACAGAGTCCTAGCAGCACA |
| AtEF1α | GCAAGATGGATGCCACTACCC | GCAAGATGGATGCCACTACCC |
| RmEXPB2 | TCTAGAATGGCTAGTTTAAGAAGCAT | CCCGGGTAGATAATTGACCACGGATC |
| RmEXPB2-qRT | TGCTGCTTGTTTGAGCAGAG | AAGCTCCACATTCCCTGCC |
| RmEXPB2-eGFP | GAGAACACGGGGGACTCTAGAATGGCTAGTTTAAGAAGCAT | GCCCTTGCTCACCATCCCGGGTAGATAATTGACCACGGATC |
基因ID Gene ID | 基因 Gene | 染色体位置 Chr location | 等电点 pI | 分子量 Molecular weight (Da) | 蛋白长度 Protein length (aa) | 亚细胞定位Subcellular localization | 信号肽长度Signal peptie length (aa) |
|---|---|---|---|---|---|---|---|
| RchiOBHmChr5g0072761 | RcEXPA1 | Chr5 | 8.12 | 27 145.26 | 254 | Cell wall | 26 |
| RchiOBHmChr7g0178811 | RcEXPA2 | Chr7 | 8.82 | 28 302.47 | 256 | Cell wall | 22 |
| RchiOBHmChr7g0199891 | RcEXPA3 | Chr7 | 9.55 | 29 064.3 | 262 | Chloroplast | 28 |
| RchiOBHmChr7g0181621 | RcEXPA4 | Chr7 | 8.64 | 26 524.79 | 250 | Cell wall | 20 |
| RchiOBHmChr6g0281211 | RcEXPA5 | Chr6 | 6.93 | 27 816.75 | 253 | Cell wall | 23 |
| RchiOBHmChr3g0488771 | RcEXPA6 | Chr3 | 7.55 | 29 302.32 | 272 | Cell wall | 24 |
| RchiOBHmChr3g0470931 | RcEXPA7 | Chr3 | 9.00 | 28 950.9 | 260 | Chloroplast | 24 |
| RchiOBHmChr4g0439541 | RcEXPA8 | Chr4 | 9.56 | 31 940.33 | 297 | Mitochondrion | |
| RchiOBHmChr3g0470941 | RcEXPA9 | Chr3 | 5.68 | 29 434.08 | 266 | Cell wall | 24 |
| RchiOBHmChr5g0033911 | RcEXPA10 | Chr5 | 9.69 | 27 795.93 | 255 | Cell wall | 30 |
| RchiOBHmChr1g0372131 | RcEXPA11 | Chr1 | 7.99 | 26 919.1 | 254 | Cell wall | 25 |
| RchiOBHmChr3g0470961 | RcEXPA12 | Chr3 | 9.68 | 31 679.04 | 286 | Chloroplast | 25 |
| RchiOBHmChr6g0245581 | RcEXPA13 | Chr6 | 5.22 | 28 332.98 | 260 | Cell wall | 27 |
| RchiOBHmChr1g0367791 | RcEXPA14 | Chr1 | 9.53 | 27 917.86 | 259 | Chloroplast | 22 |
| RchiOBHmChr3g0466461 | RcEXPA15 | Chr3 | 9.50 | 27 787.56 | 259 | Cell wall | 21 |
| RchiOBHmChr2g0166091 | RcEXPA16 | Chr2 | 8.10 | 27 554.38 | 250 | Chloroplast | 22 |
| RchiOBHmChr5g0065301 | RcEXPA17 | Chr5 | 9.68 | 27 890.68 | 260 | Cell wall | 22 |
| RchiOBHmChr3g0460691 | RcEXPA18 | Chr3 | 8.36 | 27 487.08 | 255 | Cell wall | 24 |
| RchiOBHmChr6g0303551 | RcEXPA19 | Chr6 | 8.87 | 27 306.82 | 257 | Chloroplast | 23 |
| RchiOBHmChr2g0140121 | RcEXPA20 | Chr2 | 8.39 | 35 186.7 | 313 | Cell wall | 21 |
| RchiOBHmChr2g0167701 | RcEXPA21 | Chr2 | 9.35 | 26 293.59 | 243 | Cell wall | 20 |
| RchiOBHmChr2g0166111 | RcEXPA22 | Chr2 | 6.06 | 27 581.6 | 253 | Cell wall | 23 |
| RchiOBHmChr5g0084081 | RcEXPA23 | Chr5 | 8.85 | 27 781.15 | 261 | Nucleus | 22 |
| RchiOBHmChr2g0126231 | RcEXPB1 | Chr2 | 8.73 | 27 394.3 | 255 | Cell wall | 23 |
| RchiOBHmChr1g0373921 | RcEXPB2 | Chr1 | 4.97 | 28 762.08 | 273 | Chloroplast | 23 |
| RchiOBHmChr1g0348851 | RcEXPB3 | Chr1 | 6.40 | 30 068.99 | 287 | Chloroplast | 29 |
| RchiOBHmChr6g0279781 | RcEXLA1 | Chr6 | 8.8 | 30 176.7 | 278 | Chloroplast | |
| RchiOBHmChr7g0205401 | RcEXLB1 | Chr7 | 6.87 | 27 333.67 | 250 | Cell wall | 22 |
| RchiOBHmChr7g0208241 | RcEXLB2 | Chr7 | 4.70 | 28 099.57 | 258 | Chloroplast | 25 |
Table 2 Basic information of expansin gene family members in R. chinensis'Old Blush'
基因ID Gene ID | 基因 Gene | 染色体位置 Chr location | 等电点 pI | 分子量 Molecular weight (Da) | 蛋白长度 Protein length (aa) | 亚细胞定位Subcellular localization | 信号肽长度Signal peptie length (aa) |
|---|---|---|---|---|---|---|---|
| RchiOBHmChr5g0072761 | RcEXPA1 | Chr5 | 8.12 | 27 145.26 | 254 | Cell wall | 26 |
| RchiOBHmChr7g0178811 | RcEXPA2 | Chr7 | 8.82 | 28 302.47 | 256 | Cell wall | 22 |
| RchiOBHmChr7g0199891 | RcEXPA3 | Chr7 | 9.55 | 29 064.3 | 262 | Chloroplast | 28 |
| RchiOBHmChr7g0181621 | RcEXPA4 | Chr7 | 8.64 | 26 524.79 | 250 | Cell wall | 20 |
| RchiOBHmChr6g0281211 | RcEXPA5 | Chr6 | 6.93 | 27 816.75 | 253 | Cell wall | 23 |
| RchiOBHmChr3g0488771 | RcEXPA6 | Chr3 | 7.55 | 29 302.32 | 272 | Cell wall | 24 |
| RchiOBHmChr3g0470931 | RcEXPA7 | Chr3 | 9.00 | 28 950.9 | 260 | Chloroplast | 24 |
| RchiOBHmChr4g0439541 | RcEXPA8 | Chr4 | 9.56 | 31 940.33 | 297 | Mitochondrion | |
| RchiOBHmChr3g0470941 | RcEXPA9 | Chr3 | 5.68 | 29 434.08 | 266 | Cell wall | 24 |
| RchiOBHmChr5g0033911 | RcEXPA10 | Chr5 | 9.69 | 27 795.93 | 255 | Cell wall | 30 |
| RchiOBHmChr1g0372131 | RcEXPA11 | Chr1 | 7.99 | 26 919.1 | 254 | Cell wall | 25 |
| RchiOBHmChr3g0470961 | RcEXPA12 | Chr3 | 9.68 | 31 679.04 | 286 | Chloroplast | 25 |
| RchiOBHmChr6g0245581 | RcEXPA13 | Chr6 | 5.22 | 28 332.98 | 260 | Cell wall | 27 |
| RchiOBHmChr1g0367791 | RcEXPA14 | Chr1 | 9.53 | 27 917.86 | 259 | Chloroplast | 22 |
| RchiOBHmChr3g0466461 | RcEXPA15 | Chr3 | 9.50 | 27 787.56 | 259 | Cell wall | 21 |
| RchiOBHmChr2g0166091 | RcEXPA16 | Chr2 | 8.10 | 27 554.38 | 250 | Chloroplast | 22 |
| RchiOBHmChr5g0065301 | RcEXPA17 | Chr5 | 9.68 | 27 890.68 | 260 | Cell wall | 22 |
| RchiOBHmChr3g0460691 | RcEXPA18 | Chr3 | 8.36 | 27 487.08 | 255 | Cell wall | 24 |
| RchiOBHmChr6g0303551 | RcEXPA19 | Chr6 | 8.87 | 27 306.82 | 257 | Chloroplast | 23 |
| RchiOBHmChr2g0140121 | RcEXPA20 | Chr2 | 8.39 | 35 186.7 | 313 | Cell wall | 21 |
| RchiOBHmChr2g0167701 | RcEXPA21 | Chr2 | 9.35 | 26 293.59 | 243 | Cell wall | 20 |
| RchiOBHmChr2g0166111 | RcEXPA22 | Chr2 | 6.06 | 27 581.6 | 253 | Cell wall | 23 |
| RchiOBHmChr5g0084081 | RcEXPA23 | Chr5 | 8.85 | 27 781.15 | 261 | Nucleus | 22 |
| RchiOBHmChr2g0126231 | RcEXPB1 | Chr2 | 8.73 | 27 394.3 | 255 | Cell wall | 23 |
| RchiOBHmChr1g0373921 | RcEXPB2 | Chr1 | 4.97 | 28 762.08 | 273 | Chloroplast | 23 |
| RchiOBHmChr1g0348851 | RcEXPB3 | Chr1 | 6.40 | 30 068.99 | 287 | Chloroplast | 29 |
| RchiOBHmChr6g0279781 | RcEXLA1 | Chr6 | 8.8 | 30 176.7 | 278 | Chloroplast | |
| RchiOBHmChr7g0205401 | RcEXLB1 | Chr7 | 6.87 | 27 333.67 | 250 | Cell wall | 22 |
| RchiOBHmChr7g0208241 | RcEXLB2 | Chr7 | 4.70 | 28 099.57 | 258 | Chloroplast | 25 |
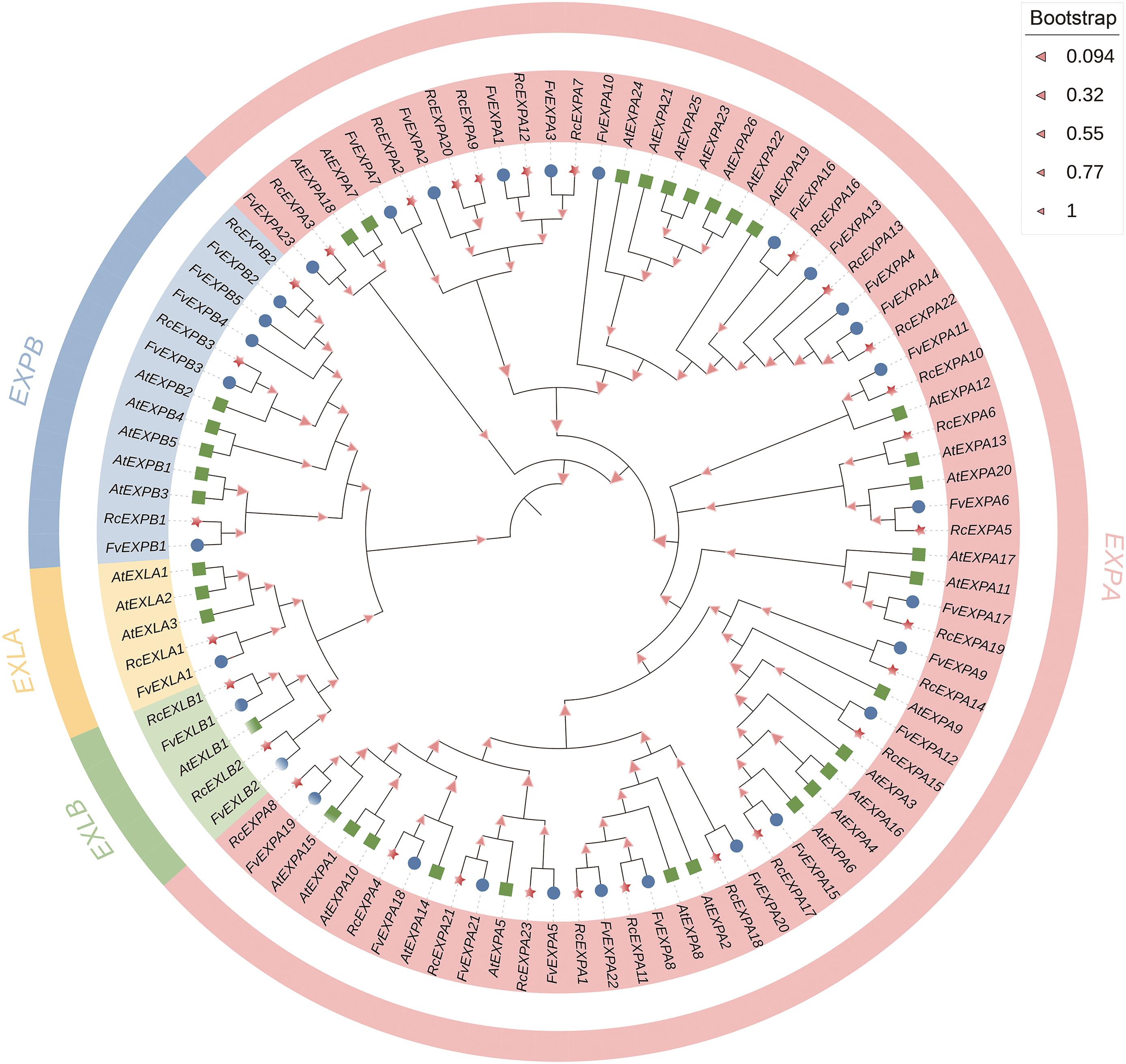
Fig. 2 Phylogenetic tree of expansin gene family in R. chinensis'Old Blush', F. avesca and ArabidopsisThe red five-pointed star indicates R.chinensis 'Old Blush', the blue circle indicates F. avesca, and the green box indicates Arabidopsis

Fig. 3 Analysis of conserved motif and gene structure of expansin gene family of R. chinensis'Old Blush'A: Conservative motif analysis. B: Conserved domain analysis. C: Gene structure analysis
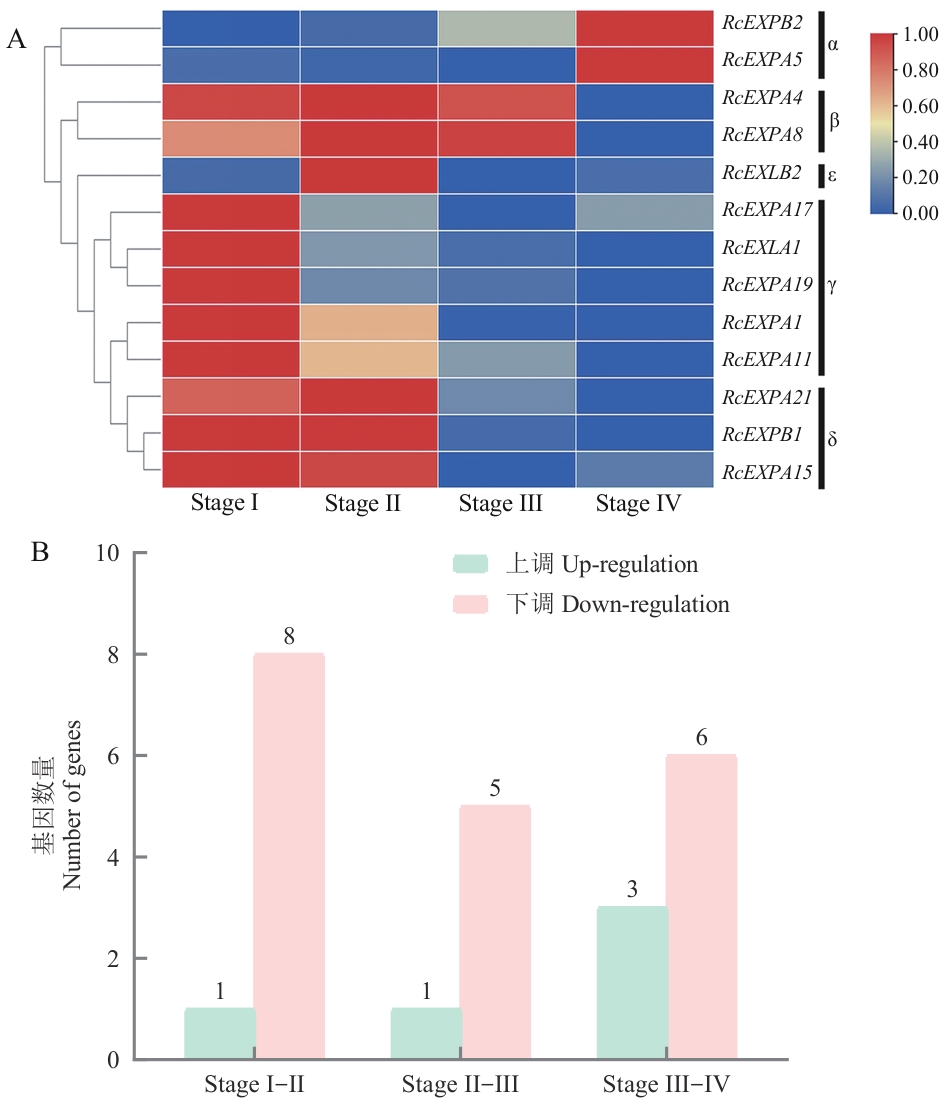
Fig. 6 Expression pattern analysis (A) and the gene number (B) of some expansin genes in different developmental stages of prickles in R. chinensis'Old Blush'Stage I: Soft and tender prickles that have not yet lignified. Stage Ⅱ: Prickles that are lignified to a certain extent and whose tips begin to bend toward the base. Stage Ⅲ: Prickles with relatively complete lignification and basically mature morphology. Stage IV: Prickles that have matured and initially begun to age

Fig. 7 Expression analysis of RmEXPB2A: Expressions of RmEXPB2 in different tissues of R. multiflora. B: Expressions of RmEXPB2 in the prickles at different developmental stages. C: RmEXPB2 expression levels in WT and RmEXPB2-OE (T1) Arabidopsis. D: RmEXPB2 expression levels in WT and RmEXPB2-OE (T2) Arabidopsis. Stage I: Starting period of R. multiflora prickles; Stage II: rapid growth period prickles; Stage III: transition period prickles; Stage IV: hardening period prickles; Stage V: maturation period prickles; line 1-10 indicates transgenic line 1‒10, WT indicates wild type. ns indicates P>0.05; * indicates P<0.05; ** indicates P<0.01; *** indicates P<0.001. The same below
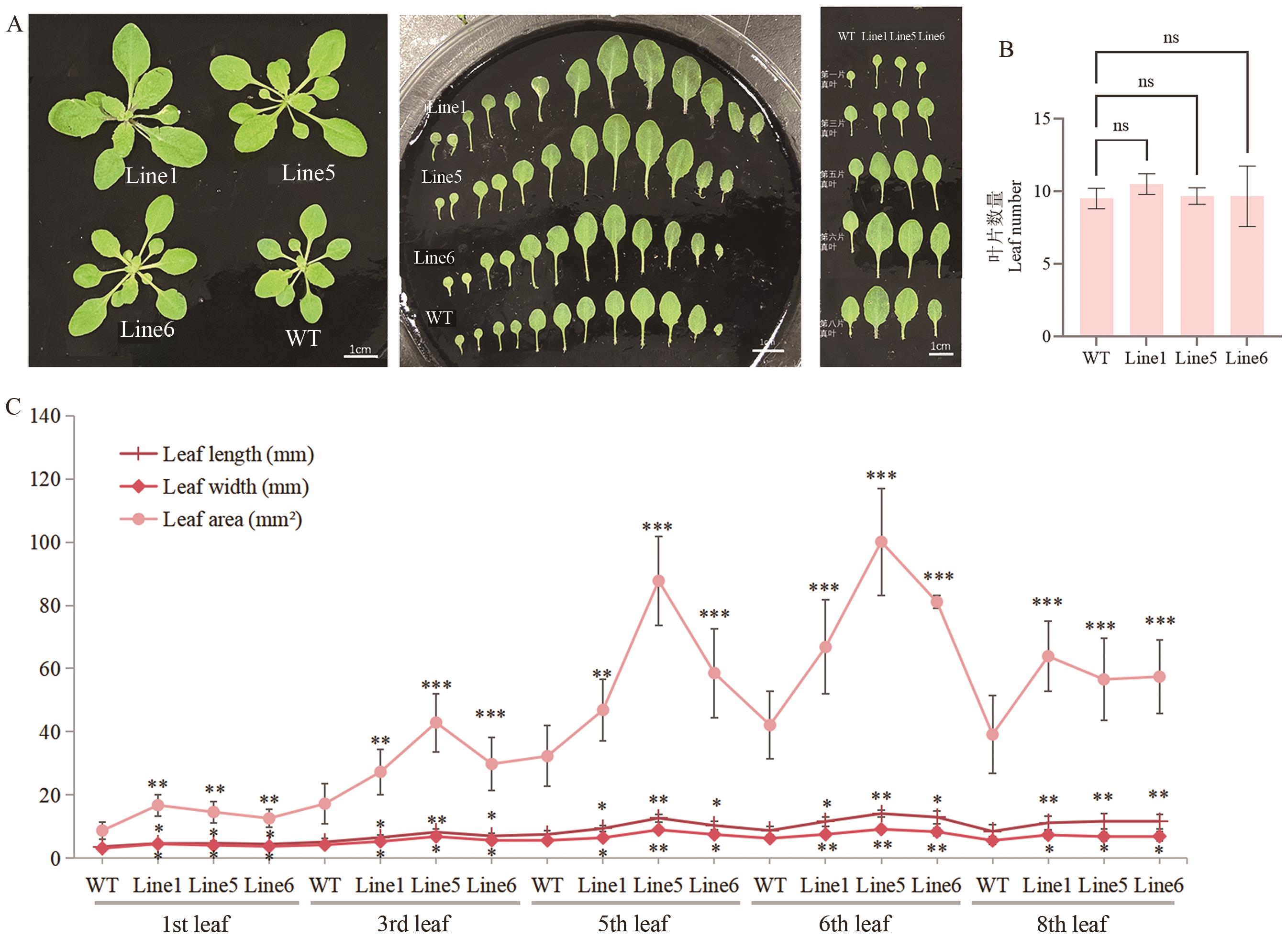
Fig. 8 Leaf phenotypes of RmEXPB2-OE (T2) Arabidopsis plantsA: True leaf phenotypic diagram. B: Number of true leaves. n>20. C: The 1st, 3rd, 5th, 6th, 8th leaf length, width and area of the true leaf. Line1, 5, 6 indicates transgenic line1, 5, 6, WT indicates wild type. The same below

Fig. 9 Root phenotypic analysis of RmEXPB2-OE (T2) Arabidopsis plantsA: Roots of WT and RmEXPB2-OE (T2) transgenic Arabidopsis. B: Root hairs of WT and RmEXPB2-OE (T2) Arabidopsis. C‒D: The main root length and hypocotyl length of WT and RmEXPB2-OE (T2) transgenic Arabidopsis. E‒F: The root hair length and root hair density of RmEXPB2-OE (T2) Arabidopsis. n>20
| [1] | Nakamura N, Hirakawa H, Sato S, et al. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses [J]. DNA Res, 2018, 25(2): 113-121. |
| [2] | Zhang Y, Zhao MJ, Zhu W, et al. Nonglandular prickle formation is associated with development and secondary metabolism-related genes in Rosa multiflora [J]. Physiol Plant, 2021, 173(3): 1147-1162. |
| [3] | McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants [J]. Plant Cell, 1992, 4(11): 1425-1433. |
| [4] | Arslan B, İncili ÇY, Ulu F, et al. Comparative genomic analysis of expansin superfamily gene members in zucchini and cucumber and their expression profiles under different abiotic stresses [J]. Physiol Mol Biol Plants, 2021, 27(12): 2739-2756. |
| [5] | Cosgrove DJ. Plant cell wall loosening by expansins [J]. Annu Rev Cell Dev Biol, 2024, 40(1): 329-352. |
| [6] | Wang ZZ, Cao JB, Lin N, et al. Origin, evolution, and diversification of the expansin family in plants [J]. Int J Mol Sci, 2024, 25(21): 11814. |
| [7] | Kök BÖ, Celik Altunoglu Y, Öncül AB, et al. Expansin gene family database: a comprehensive bioinformatics resource for plant expansin multigene family [J]. J Bioinform Comput Biol, 2023, 21(3): 2350015. |
| [8] | Liu WM, Lyu TQ, Xu LA, et al. Complex molecular evolution and expression of expansin gene families in three basic diploid species of Brassica [J]. Int J Mol Sci, 2020, 21(10): 3424. |
| [9] | Goh HH, Sloan J, Dorca-Fornell C, et al. Inducible repression of multiple expansin genes leads to growth suppression during leaf development [J]. Plant Physiol, 2012, 159(4): 1759-1770. |
| [10] | Yan A, Wu MJ, Yan LM, et al. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis [J]. PLoS One, 2014, 9(1): e85208. |
| [11] | Feng X, Li CT, He FM, et al. Genome-wide identification of expansin genes in wild soybean (Glycine soja) and functional characterization of Expansin B1 (GsEXPB1) in soybean hair root [J]. Int J Mol Sci, 2022, 23(10): 5407. |
| [12] | Liu WM, Xu LA, Lin H, et al. Two expansin genes, AtEXPA4 and AtEXPB5, are redundantly required for pollen tube growth and AtEXPA4 is involved in primary root elongation in Arabidopsis thaliana [J]. Genes, 2021, 12(2): 249. |
| [13] | Jiang FL, Lopez A, Jeon S, et al. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking [J]. Hortic Res, 2019, 6: 17. |
| [14] | Calderini DF, Castillo FM, Arenas-M A, et al. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential [J]. New Phytol, 2021, 230(2): 629-640. |
| [15] | Dai FW, Zhang CQ, Jiang XQ, et al. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals [J]. Plant Physiol, 2012, 160(4): 2064-2082. |
| [16] | Lü PT, Kang M, Jiang XQ, et al. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis [J]. Planta, 2013, 237(6): 1547-1559. |
| [17] | Raymond O, Gouzy J, Just J, et al. The Rosagenome provides new insights into the domestication of modern roses [J]. Nat Genet, 2018, 50(6): 772-777. |
| [18] | Han Y, Yu JY, Zhao T, et al. Dissecting the genome-wide evolution and function of R2R3-MYB transcription factor family in Rosa chinensis [J]. Genes, 2019, 10(10): 823. |
| [19] | 由玉婉, 张雨, 孙嘉毅, 等. ‘月月粉’月季NAC家族全基因组鉴定及皮刺发育相关成员的筛选 [J]. 中国农业科学, 2022, 55(24): 4895-4911. |
| You YW, Zhang Y, Sun JY, et al. Genome-wide identification of NAC family and screening of its members related to prickle development in Rosa chinensis old blush [J]. Sci Agric Sin, 2022, 55(24): 4895-4911. | |
| [20] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [21] | Hepler NK, Bowman A, Carey RE, et al. Expansin gene loss is a common occurrence during adaptation to an aquatic environment [J]. Plant J, 2020, 101(3): 666-680. |
| [22] | Krishnamurthy P, Hong JK, Kim JA, et al. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication [J]. Mol Genet Genomics, 2015, 290(2): 521-530. |
| [23] | Liu XP, Dong SY, Miao H, et al. Genome-wide analysis of expansins and their role in fruit spine development in cucumber (Cucumis sativus L.) [J]. Hortic Plant J, 2022, 8(6): 757-768. |
| [24] | Sampedro J, Carey RE, Cosgrove DJ. Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs [J]. J Plant Res, 2006, 119(1): 11-21. |
| [25] | Zhang W, Yan HW, Chen WJ, et al. Genome-wide identification and characterization of maize expansin genes expressed in endosperm [J]. Mol Genet Genomics, 2014, 289(6): 1061-1074. |
| [26] | Qiu DY, Xu SL, Wang Y, et al. Primary cell wall modifying proteins regulate wall mechanics to steer plant morphogenesis [J]. Front Plant Sci, 2021, 12: 751372. |
| [27] | Ono K. Signal peptides and their fragments in post-translation: novel insights of signal peptides [J]. Int J Mol Sci, 2024, 25(24): 13534. |
| [28] | Zhang JL, Dong TT, Zhu MK, et al. Transcriptome- and genome-wide systematic identification of expansin gene family and their expression in tuberous root development and stress responses in sweetpotato (Ipomoea batatas) [J]. Front Plant Sci, 2024, 15: 1412540. |
| [29] | Lou Y, Zhou HS, Han Y, et al. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana [J]. New Phytol, 2018, 217(1): 378-391. |
| [30] | Zhou NN, Simonneau F, Thouroude T, et al. Morphological studies of rose prickles provide new insights [J]. Hortic Res, 2021, 8(1): 221. |
| [31] | Swarnkar MK, Kumar P, Dogra V, et al. Prickle morphogenesis in rose is coupled with secondary metabolite accumulation and governed by canonical MBW transcriptional complex [J]. Plant Direct, 2021, 5(6): e00325. |
| [32] | Saint-Oyant LH, Ruttink T, Hamama L, et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits [J]. Nat Plants, 2018, 4(7): 473-484. |
| [33] | Xing SC, Li F, Guo QF, et al. The involvement of an expansin gene TaEXPB23 from wheat in regulating plant cell growth [J]. Biol Plant, 2009, 53(3): 429-434. |
| [34] | Zou HY, Wenwen YH, Zang GC, et al. OsEXPB2, a β-expansin gene, is involved in rice root system architecture [J]. Mol Breed, 2015, 35(1): 41. |
| [35] | Lin CF, Choi HS, Cho HT. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis [J]. Mol Cells, 2011, 31(4): 393-398. |
| [36] | Chen SK, Luo YX, Wang GJ, et al. Genome-wide identification of expansin genes in Brachypodium distachyon and functional characterization of BdEXPA27 [J]. Plant Sci, 2020, 296: 110490. |
| [1] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [2] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [3] | HUANG Xu-sheng, ZHOU Ya-li, CHAI Xu-dong, WEN Jing, WANG Ji-ping, JIA Xiao-yun, LI Run-zhi. Cloning of Plastidial PfLPAT1B Gene from Perilla frutescens and Its Functional Analysis in Oil Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(7): 226-236. |
| [4] | SONG Jia-yi, SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen. Identification of the Snapdragon Expansin Gene Family and Screening of Its Genes Related to Resistance to Sclerotinia sclerotiorum [J]. Biotechnology Bulletin, 2025, 41(4): 227-242. |
| [5] | LI Bin, SU Xiang-ping, LIU Chang, WANG Yu-bing, ZHANG Yong-hong, ZHOU Chao, XU Qing. Chloroplast Genome Characteristics and Phylogenetic Analysis of Scrophulariaceae [J]. Biotechnology Bulletin, 2025, 41(3): 240-254. |
| [6] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [7] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [8] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [9] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [10] | YIN Liang, WANG Dai-wei, LIU Yue-ying, LIU Hai-yan, LUO Guang-hong. Cloning and Expression of Protease SpP1 Gene and Characterization of Enzymatic Properties [J]. Biotechnology Bulletin, 2024, 40(4): 278-286. |
| [11] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [12] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [13] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [14] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [15] | CHEN Nan-nan, WANG Chun-lai, JIANG Zhen-zhong, JIAO Peng, GUAN Shu-yan, MA Yi-yong. Genetic Transformation and Chilling Resistance Analysis of Maize ZmDHN15 Gene in Tobacco [J]. Biotechnology Bulletin, 2023, 39(4): 259-267. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||