








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 150-160.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0598
Previous Articles Next Articles
YANG Juan( ), FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei(
), FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei( )
)
Received:2025-06-10
Online:2026-01-26
Published:2026-02-04
Contact:
ZHAO Shi-wei
E-mail:yangjuanyl@126.com;2668587780@qq.com
YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose[J]. Biotechnology Bulletin, 2026, 42(1): 150-160.

Fig. 1 Different flower development stages of 'Lady of Shalott'S1: Sepals tightly wrapped, petals not colored. S2: Sepals open, petals begin to color. S3: Sepals fully open, petals fully colored. S4: Petals fully bloomed. The same below
| 用途 Application | 引物名称Primer name | 引物序列Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene cloning | RcERF4-F1 | ATGGCACCGAGAACCGAGA |
| RcERF4-R1 | TCAGGCGAGTTCCGCCGGA | |
| RcRAP2-12-F1 | ATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R1 | GAAAACTCCCCCAGACATTG | |
实时荧光定量PCR Quantitative real-time PCR | RcERF4-F2 | ACCACGGCTAAACCTAACGG |
| RcERF4-R2 | ATTCACATTCTCCGACGGCA | |
| RcRAP2-12-F2 | CGACTTGGTCTGGCCTGATT | |
| RcRAP2-12-R2 | TGCACTCCACGGATTTCACA | |
亚细胞定位和遗传转化烟草 Subcellular localization and genetic transformation of tobacco | RcERF4-F3 | aagtccggagctagctctagaATGGCACCGAGAACCGAGA |
| RcERF4-R3 | gcccttgctcaccatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F3 | aagtccggagctagctctagaATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R3 | gcccttgctcaccatggatccGAAAACTCCCCCAGACATTGAA | |
酵母双杂交 Yeast two-hybrid | RcERF4-F4 | gccatggaggccagtgaattcATGGCACCGAGAACCGAGA |
| RcERF4-R4 | cagctcgagctcgatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F4 | tcagaggaggacctgcatatgATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R4 | ctagttatgcggccgctgcagGAAAACTCCCCCAGACATTGAA | |
双分子荧光互补 Bimolecular fluorescence complementation | RcERF4-F5 | tggcgcgccactagtggatccATGGCACCGAGAACCGAGA |
| RcERF4-R5 | ctccatcccgggagcggtaccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F5 | tggcgcgccactagtggatccATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R5 | gtacatcccgggagcggtaccGAAAACTCCCCCAGACATTGAA |
Table 1 Primers used in this study
| 用途 Application | 引物名称Primer name | 引物序列Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene cloning | RcERF4-F1 | ATGGCACCGAGAACCGAGA |
| RcERF4-R1 | TCAGGCGAGTTCCGCCGGA | |
| RcRAP2-12-F1 | ATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R1 | GAAAACTCCCCCAGACATTG | |
实时荧光定量PCR Quantitative real-time PCR | RcERF4-F2 | ACCACGGCTAAACCTAACGG |
| RcERF4-R2 | ATTCACATTCTCCGACGGCA | |
| RcRAP2-12-F2 | CGACTTGGTCTGGCCTGATT | |
| RcRAP2-12-R2 | TGCACTCCACGGATTTCACA | |
亚细胞定位和遗传转化烟草 Subcellular localization and genetic transformation of tobacco | RcERF4-F3 | aagtccggagctagctctagaATGGCACCGAGAACCGAGA |
| RcERF4-R3 | gcccttgctcaccatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F3 | aagtccggagctagctctagaATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R3 | gcccttgctcaccatggatccGAAAACTCCCCCAGACATTGAA | |
酵母双杂交 Yeast two-hybrid | RcERF4-F4 | gccatggaggccagtgaattcATGGCACCGAGAACCGAGA |
| RcERF4-R4 | cagctcgagctcgatggatccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F4 | tcagaggaggacctgcatatgATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R4 | ctagttatgcggccgctgcagGAAAACTCCCCCAGACATTGAA | |
双分子荧光互补 Bimolecular fluorescence complementation | RcERF4-F5 | tggcgcgccactagtggatccATGGCACCGAGAACCGAGA |
| RcERF4-R5 | ctccatcccgggagcggtaccGGCGAGTTCCGCCGGAGG | |
| RcRAP2-12-F5 | tggcgcgccactagtggatccATGTGCGGAGGTGCCATTAT | |
| RcRAP2-12-R5 | gtacatcccgggagcggtaccGAAAACTCCCCCAGACATTGAA |
| 名称 Name | 网址 Website | 用途 Application |
|---|---|---|
| ExPASy-ProtParam | https://web.expasy.org/protparam/ | 理化性质 Physicochemical properties |
| CDS | https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 结构域 Domain |
| TMHMM-2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | 跨膜结构 Transmembrane structure |
| SignalP-4.1 | https://services.healthtech.dtu.dk/services/SignalP-4.1/ | 信号肽 Signal peptide |
| NetGlycate-1.0 | https://services.healthtech.dtu.dk/services/NetGlycate-1.0/ | 糖基化位点 Glycosylation site |
| NetPhos-3.1 | https://services.healthtech.dtu.dk/services/NetPhos-3.1/ | 磷酸化位点 Phosphorylation site |
Table 2 Bioinformatics analysis websites and their applications
| 名称 Name | 网址 Website | 用途 Application |
|---|---|---|
| ExPASy-ProtParam | https://web.expasy.org/protparam/ | 理化性质 Physicochemical properties |
| CDS | https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 结构域 Domain |
| TMHMM-2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | 跨膜结构 Transmembrane structure |
| SignalP-4.1 | https://services.healthtech.dtu.dk/services/SignalP-4.1/ | 信号肽 Signal peptide |
| NetGlycate-1.0 | https://services.healthtech.dtu.dk/services/NetGlycate-1.0/ | 糖基化位点 Glycosylation site |
| NetPhos-3.1 | https://services.healthtech.dtu.dk/services/NetPhos-3.1/ | 磷酸化位点 Phosphorylation site |

Fig. 4 Amino acid sequence alignment of RcERF4 (A) and RcRAP2-12 (B)Aa: Argentina anserina, XP_050371303.1. Ej: Eriobotrya japonica, AKN10304.1. Fa: Fragaria × ananassa, AZL19463.1. Fv: Fragaria vesca subsp. vesca, XP_004293846.1. Mp: Malus pumila, NP_001385262.1. Ms: Malus sylvestris, XP_050108098.1. Pp: Prunus persica, ALO81027.1. Ra: Rubus argutus, KAK9936415.1. Rc: Rosa chinensis, XP_024158160.1. Rr: Rosa rugosa, XP_062013106.1. Pa: Prunus avium, XP_021819942.1. Pb: Pyrus × bretschneideri, XP_009338318.1. Ps: Prunus serrulata, APY26916.1. The red dot indicates the WLG component. The red square indicates the target protein. The red box indicates the 14th and 19th amino acids. The blue box indicates the AP2 domain
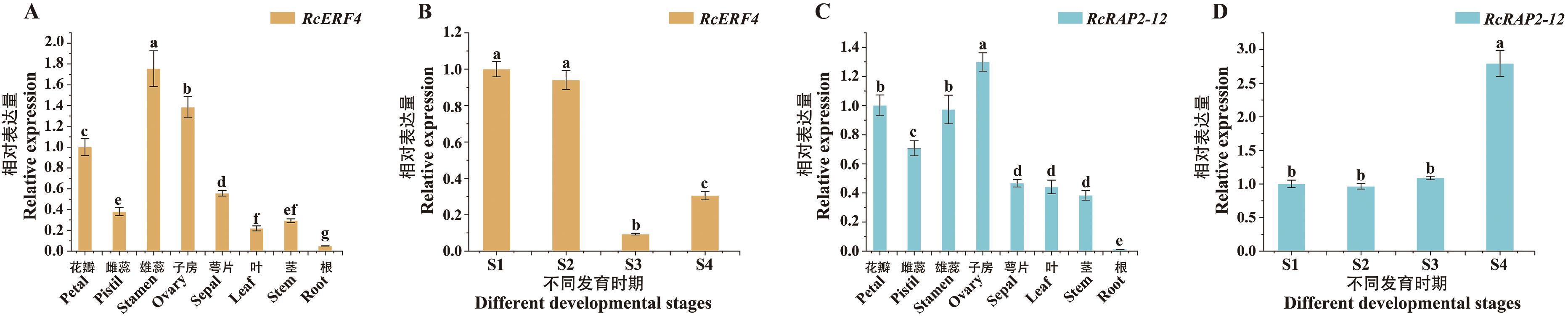
Fig. 6 Expression patterns of RcERF4 and RcRAP2-12 in different tissues and various floral developmental stages of 'Lady of Shalott'Three biological replicates were used. The error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA, different lower case letters indicate significant differences (P<0.05), the same below
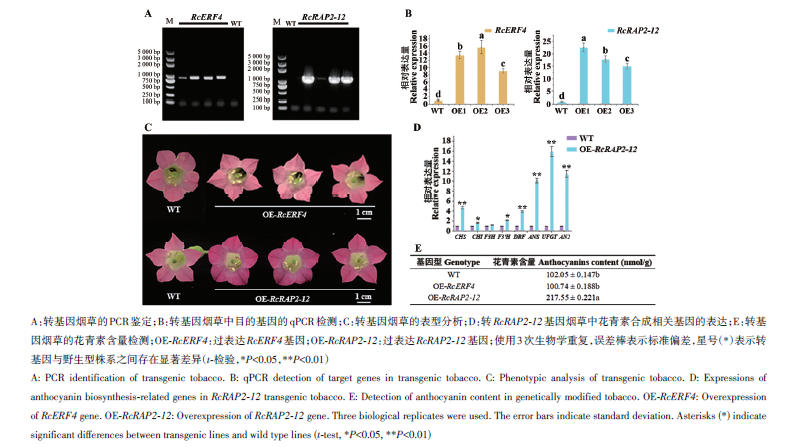
Fig. 8 Identification and analysis of transgenic tobaccoA: PCR identification of transgenic tobacco. B: qPCR detection of target genes in transgenic tobacco. C: Phenotypic analysis of transgenic tobacco. D: Expressions of anthocyanin biosynthesis-related genes in RcRAP2-12 transgenic tobacco. E: Detection of anthocyanin content in genetically modified tobacco. OE-RcERF4: Overexpression of RcERF4 gene. OE-RcRAP2-12: Overexpression of RcRAP2-12 gene. Three biological replicates were used. The error bars indicate standard deviation. Asterisks (*) indicate significant differences between transgenic lines and wild type lines (t-test, *P<0.05, **P<0.01)
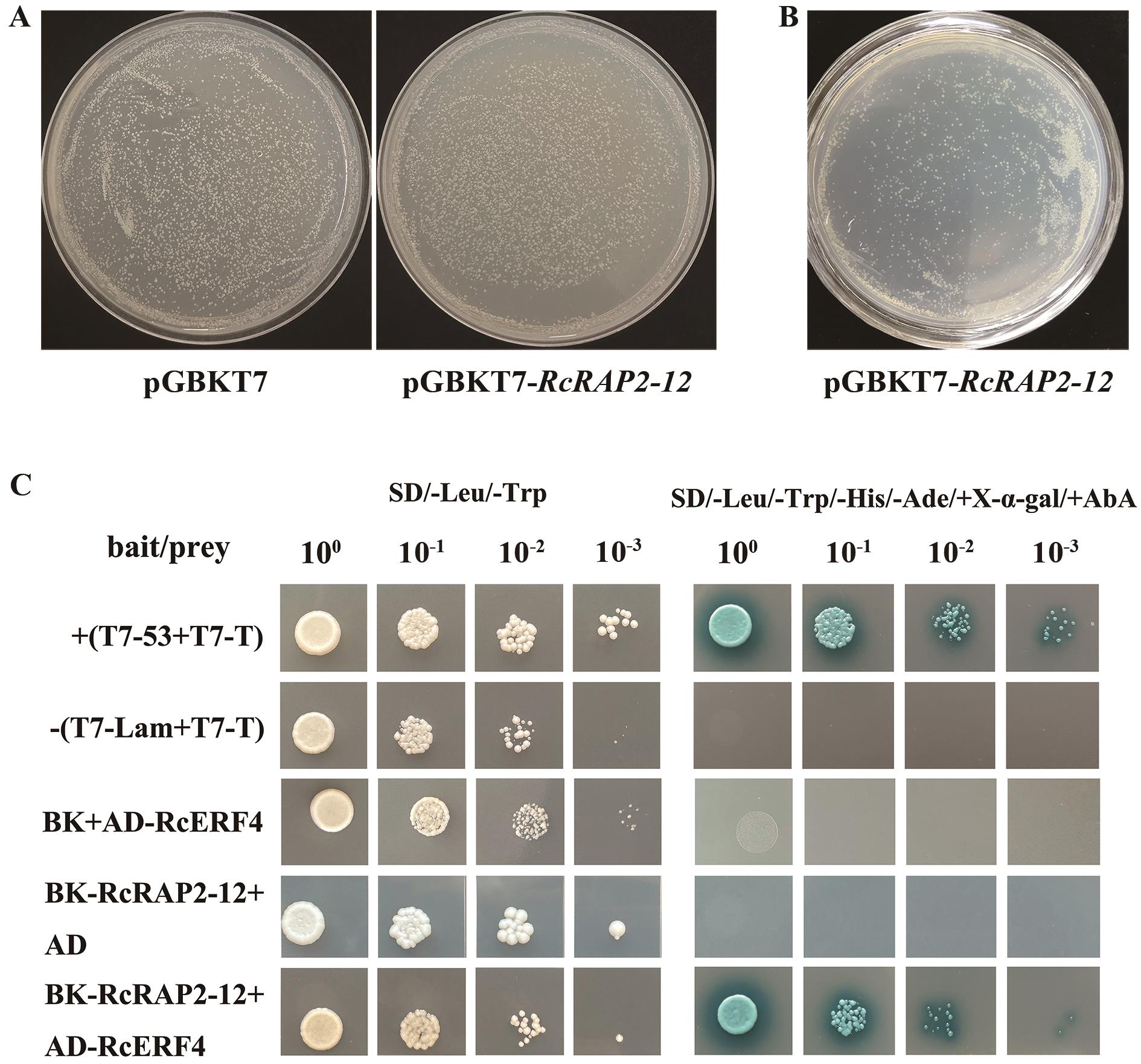
Fig. 9 Yeast two-hybrid verificationA: Toxicity testing of pGBKT7-RAP2-12. B: Self-activation verification of pGBKT7-RAP2-12. C: Interaction validation between pGBKT7-RAP2-12 and pGADT7-RcERF4
| [1] | Ma ZM, Jin YM, Wu T, et al. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice [J]. Front Plant Sci, 2022, 13: 1007811. |
| [2] | Li HY, Du HP, Huang ZR, et al. The AP2/ERF transcription factor TOE4b regulates photoperiodic flowering and grain yield per plant in soybean [J]. Plant Biotechnol J, 2023, 21(8): 1682-1694. |
| [3] | Shi HY, Qanmber G, Yang ZR, et al. An AP2/ERF transcription factor GhERF109 negatively regulates plant growth and development in cotton [J]. Plant Sci, 2025, 352: 112365. |
| [4] | Zhang Y, Ji AJ, Xu ZC, et al. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza [J]. Plant Mol Biol, 2019, 100(1/2): 83-93. |
| [5] | Zhang J, Liu ZY, Sakamoto S, et al. ETHYLENE RESPONSE FACTOR 34 promotes secondary cell wall thickening and strength of rice peduncles [J]. Plant Physiol, 2022, 190(3): 1806-1820. |
| [6] | Xu L, Yang LJ, Li AP, et al. An AP2/ERF transcription factor confers chilling tolerance in rice [J]. Sci Adv, 2024, 10(35): eado4788. |
| [7] | Ritonga FN, Ngatia JN, Wang YR, et al. AP2/ERF, an important cold stress-related transcription factor family in plants: a review [J]. Physiol Mol Biol Plants, 2021, 27(9): 1953-1968. |
| [8] | Su ZL, Li AM, Wang M, et al. The role of AP2/ERF transcription factors in plant responses to biotic stress [J]. Int J Mol Sci, 2025, 26(10): 4921. |
| [9] | 兰孟焦, 后猛, 肖满秋, 等. AP2/ERF转录因子参与植物次生代谢和逆境胁迫响应的研究进展 [J]. 植物遗传资源学报, 2023, 24(5): 1223-1235. |
| Lan MJ, Kou M, Xiao MQ, et al. Research progress of AP2/ERF transcription factors participating in plant secondary metabolism and stress response [J]. J Plant Genet Resour, 2023, 24(5): 1223-1235. | |
| [10] | 卢勇杰, 夏海乾, 李永铃, 等. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析 [J]. 生物技术通报, 2025, 41(4): 266-277. |
| Lu YJ, Xia HQ, Li YL, et al. Cloning and expression analysis of AP2/ERF transcription factor NtESR2 in Nicotiana tabacum [J]. Biotechnol Bull, 2025, 41(4): 266-277. | |
| [11] | Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression [J]. Biochem Biophys Res Commun, 2002, 290(3): 998-1009. |
| [12] | Khaskheli AJ, Ahmed W, Ma C, et al. RhERF113 functions in ethylene-induced petal senescence by modulating cytokinin content in rose [J]. Plant Cell Physiol, 2018, 59(12): 2442-2451. |
| [13] | An JP, Zhang XW, Bi SQ, et al. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple [J]. Plant J, 2020, 101(3): 573-589. |
| [14] | Zhang SY, Liu S, Ren YS, et al. The ERF transcription factor ZbERF3 promotes ethylene-induced anthocyanin biosynthesis in Zanthoxylum bungeanum [J]. Plant Sci, 2024, 349: 112264. |
| [15] | Mo RL, Han GM, Zhu ZX, et al. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘Zijin’ mulberry fruits by interacting with MYBA and F3H genes [J]. Int J Mol Sci, 2022, 23(14): 7615. |
| [16] | Cao YW, Song M, Bi MM, et al. Lily (Lilium spp.) LhERF4 negatively affects anthocyanin biosynthesis by suppressing LhMYBSPLATTER transcription [J]. Plant Sci, 2024, 342: 112026. |
| [17] | Sun HY, Hu KD, Wei SW, et al. ETHYLENE RESPONSE FACTORS 4.1/4.2 with an EAR motif repress anthocyanin biosynthesis in red-skinned pears [J]. Plant Physiol, 2023, 192(3): 1892-1912. |
| [18] | Jiang SH, Wang HH, Zhang R, et al. Transcriptomic-based analysis to identify candidate genes for blue color rose breeding [J]. Plant Mol Biol, 2023, 111(4/5): 439-454. |
| [19] | Su MY, Damaris RN, Hu ZR, et al. Metabolomic analysis on the petal of ‘Chen Xi’ rose with light-induced color changes [J]. Plants, 2021, 10(10): 2065. |
| [20] | Ji NZ, Wang QY, Li SS, et al. Metabolic profile and transcriptome reveal the mystery of petal blotch formation in rose [J]. BMC Plant Biol, 2023, 23(1): 46. |
| [21] | 王超林. RcGT1和Rc3GGT在月季粉色系花瓣呈色过程中的功能研究 [D]. 北京: 北京农学院, 2023. |
| Wang CL. Function of RcGT1 and Rc3GGT for petal coloration in pink Rosa chinensis [D]. Beijing: Beijing University Of Agriculture, 2023. | |
| [22] | Yan HJ, Zhang H, Wang QG, et al. The Rosa chinensis cv. viridiflora phyllody phenotype is associated with misexpression of flower organ identity genes [J]. Front Plant Sci, 2016, 7: 996. |
| [23] | Gallois P, Marinho P. Leaf disk transformation using Agrobacterium tumefaciens -expression of heterologous genes in tobacco [J]. Methods Mol Biol, 1995, 49: 39-48. |
| [24] | 杨娟. 基于多组学分析的燕子花花色变异分子机理 [D]. 哈尔滨: 东北林业大学, 2023. |
| Yang J. The molecular mechanism of flower color variation of iris laevigata based on multi-omics analysis [D]. Harbin: Northeast Forestry University, 2023. | |
| [25] | Wang Z, Song GW, Zhang FJ, et al. Functional characterization of AP2/ERF transcription factors during flower development and anthocyanin biosynthesis related candidate genes in Lycoris [J]. Int J Mol Sci, 2023, 24(19): 14464. |
| [26] | Li DL, He YJ, Li SH, et al. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.) [J]. Plant Physiol Biochem, 2021, 167: 492-503. |
| [27] | Wang Z, Yang JC, Gao Q, et al. The transcription factor NtERF13a enhances abiotic stress tolerance and phenylpropanoid compounds biosynthesis in tobacco [J]. Plant Sci, 2023, 334: 111772. |
| [28] | He GR, Zhang R, Jiang SH, et al. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis [J]. Hortic Res, 2023, 10(6): uhad080. |
| [29] | Wang YG, Zhou LJ, Wang YX, et al. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of Chrysanthemum [J]. Sci Hortic, 2022, 293: 110674. |
| [30] | Zhou LJ, Wang YX, Wang YG, et al. Transcription factor CmbHLH16 regulates petal anthocyanin homeostasis under different lights in Chrysanthemum [J]. Plant Physiol, 2022, 190(2): 1134-1152. |
| [31] | Ji XL, Zhao LL, Liu BY, et al. MdZFP7 integrates JA and GA signals via interaction with MdJAZ2 and MdRGL3a in regulating anthocyanin biosynthesis and undergoes degradation by the E3 ubiquitin ligase MdBRG3 [J]. J Integr Plant Biol, 2025, 67(5): 1339-1363. |
| [1] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [2] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [3] | ZHANG Yue, DAI Yue-hua, ZHANG Ying-ying, LI Ao-hui, LI Chu-hui, XUE Jin-ai, QIN Hui-bin, CHEN Yan, NIE Meng-en, ZHANG Hai-ping. Cloning and Functional Analysis of the Soybean Enoyl-CoA Reductase ECR14 Gene [J]. Biotechnology Bulletin, 2026, 42(1): 95-104. |
| [4] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [5] | ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L. [J]. Biotechnology Bulletin, 2026, 42(1): 208-217. |
| [6] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| [7] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [8] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [9] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [10] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [11] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [12] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [13] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [14] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [15] | HU Lu, WANG Kai, XU Jing-yi, YE Li-hui, WANG Yong-fei, WANG Li-hua, LI Jie-qin. Research Progress in Genetic Transformation Technologies of Maize and Sorghum [J]. Biotechnology Bulletin, 2025, 41(9): 32-43. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||