








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 67-75.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0838
Previous Articles Next Articles
LIU Yu1,2( ), LIU Xiao-xiao2,3, ZHENG Jian-hao2,4, LI Bin2, HE Hong-hong1(
), LIU Xiao-xiao2,3, ZHENG Jian-hao2,4, LI Bin2, HE Hong-hong1( ), MAO Li1,2(
), MAO Li1,2( )
)
Received:2025-08-02
Online:2026-01-26
Published:2026-02-04
Contact:
HE Hong-hong, MAO Li
E-mail:Y18728628336@outlook.com;honghong3h@126.com;mao-li@live.cn
LIU Yu, LIU Xiao-xiao, ZHENG Jian-hao, LI Bin, HE Hong-hong, MAO Li. Preparation and Identification of Monoclonal Antibodies against the Bovine Coronavirus[J]. Biotechnology Bulletin, 2026, 42(1): 67-75.
引物名称 Primer | 寡核苷酸序列 Oligonucleotide sequence(5'-3') | 基因长度 Gene length (bp) |
|---|---|---|
| cpCoV-S-F | ATGTTTTTGATACTTTTAATTTC | 4 092 |
| cpCoV-S-R | GTCGTCATGTGAGGTTTT | |
| cpCoV-S1-F | ATGTTTTTGATACTTTTAATTTC | 2 304 |
| cpCoV-S1-R | TCTACGACTTCGTCTTTTTG | |
| cpCoV-S2-F | AGACGAAGTCGTAGATCG | 1 803 |
| cpCoV-S2-R | GTCGTCATGTGAGGTTTT |
Table 1 Primer sequences
引物名称 Primer | 寡核苷酸序列 Oligonucleotide sequence(5'-3') | 基因长度 Gene length (bp) |
|---|---|---|
| cpCoV-S-F | ATGTTTTTGATACTTTTAATTTC | 4 092 |
| cpCoV-S-R | GTCGTCATGTGAGGTTTT | |
| cpCoV-S1-F | ATGTTTTTGATACTTTTAATTTC | 2 304 |
| cpCoV-S1-R | TCTACGACTTCGTCTTTTTG | |
| cpCoV-S2-F | AGACGAAGTCGTAGATCG | 1 803 |
| cpCoV-S2-R | GTCGTCATGTGAGGTTTT |
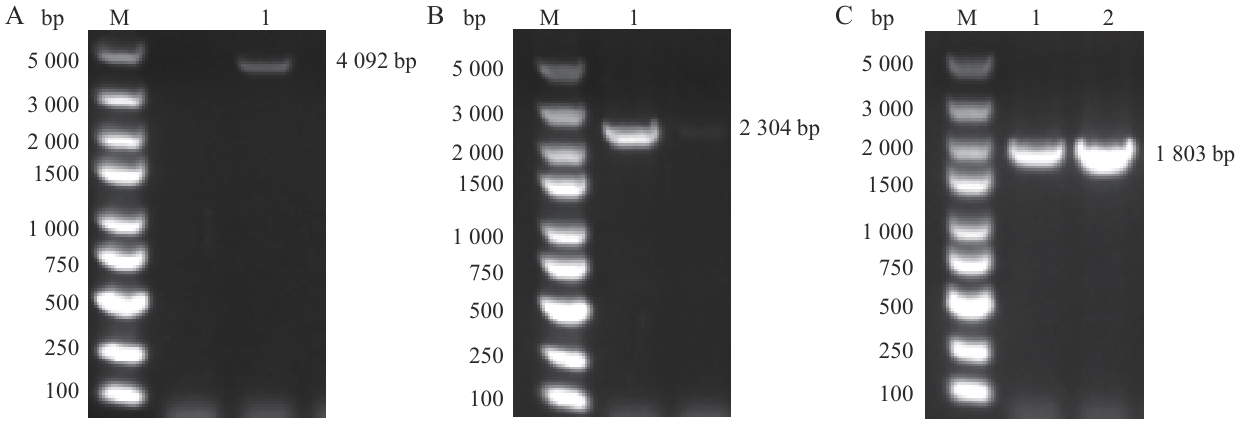
Fig. 3 Identification results of plasmids of S geneA: PCR amplification of pCAGGS-cpCoV-S-HA. B: PCR amplification of pCAGGS-cpCoV-S1-HA. C: PCR amplification of pCAGGS-cpCoV-S2-HA. M: DNA molecular weight standard. 1, 2: RT-PCR products of the target gene
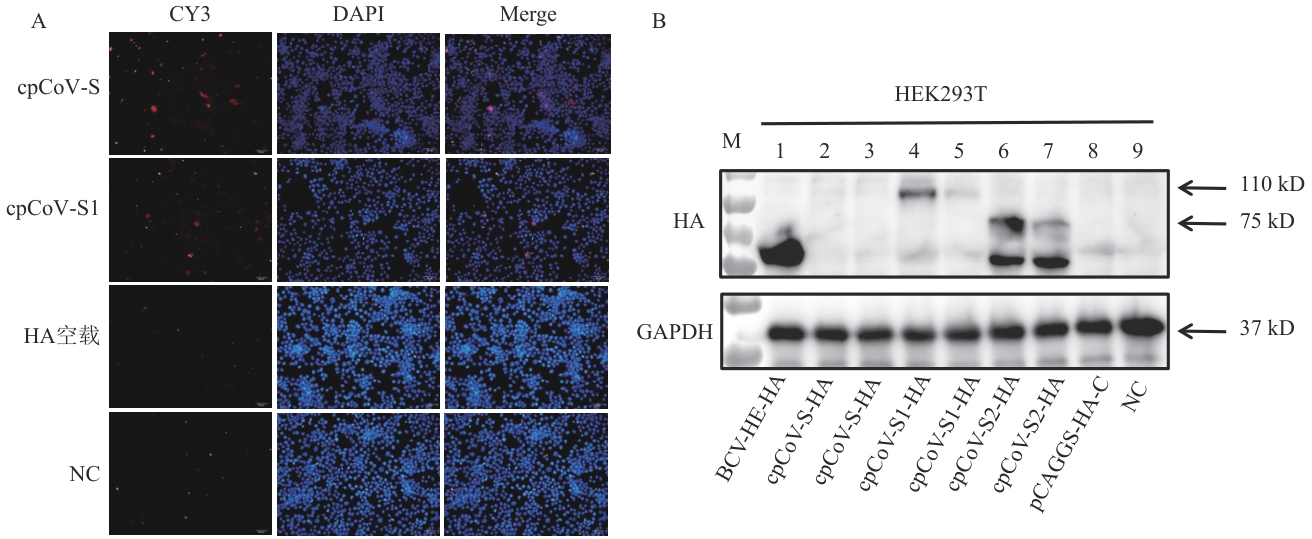
Fig. 4 Expression and identification of S proteinA: IFA test results. B: Western-blot test results. M: Protein molecular weight standard. 1: Positive plasmid control of HA-BCoV-HE. 2, 3: Western-blot test results of HA-cpCoV-S recombinant plasmid. 4, 5: Western-blot test results of HA-cpCoV-S1 recombinant plasmid. 6, 7: Western-blot test results of HA-cpCoV-S2 recombinant plasmid. 8: Empty pCAGGS-HA; 9: 293T cells control
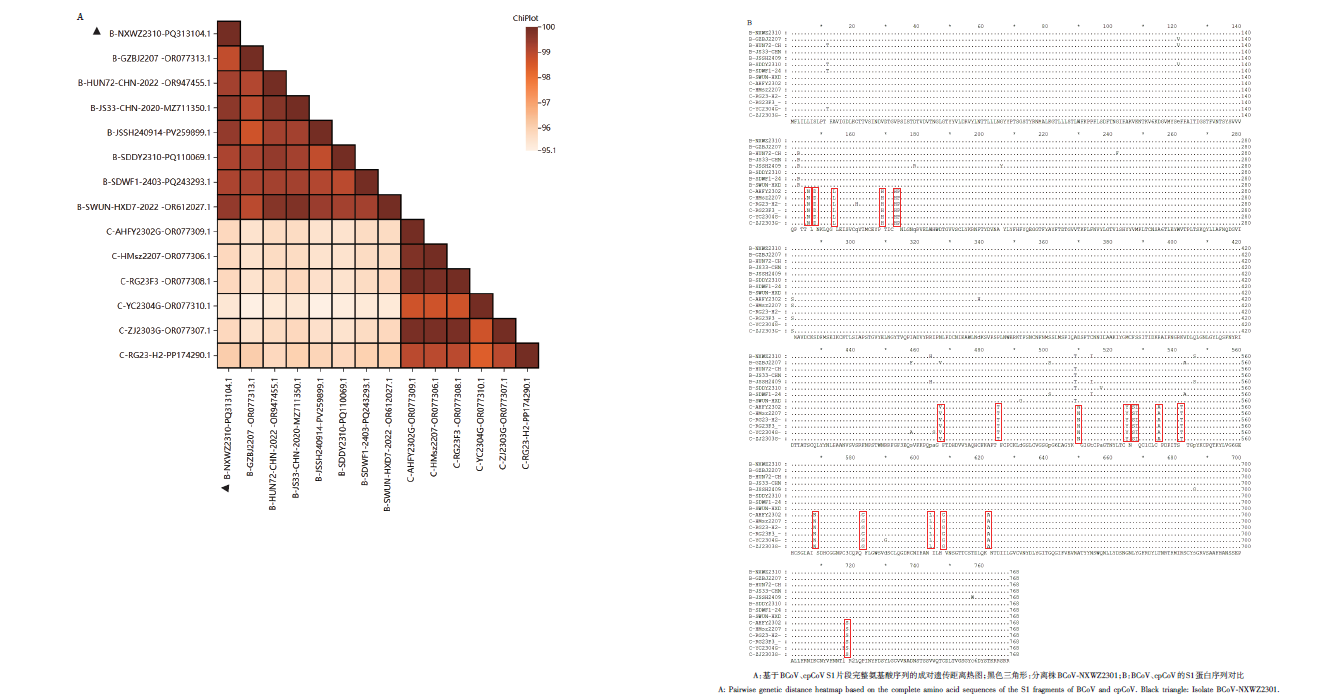
Fig. 6 Analysis results of homology and differential proteins of BCoV and cpCoV S1 proteinsA: Pairwise genetic distance heatmap based on the complete amino acid sequences of the S1 fragments of BCoV and cpCoV. Black triangle: Isolate BCoV-NXWZ2301. B: Comparison of the S1 protein sequences of BCoV and cpCoV
| [1] | Holmes EC. The emergence and evolution of SARS-CoV-2 [J]. Annu Rev Virol, 2024, 11: 21-42. |
| [2] | Woo PCY, Lau SKP, Lam CSF, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus [J]. J Virol, 2012, 86(7): 3995-4008. |
| [3] | Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses [J]. Nat Rev Microbiol, 2019, 17(3): 181-192. |
| [4] | Zhong N, Zheng B, Li Y, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003 [J]. Lancet, 2003, 362(9393): 1353-1358. |
| [5] | Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia [J]. N Engl J Med, 2012, 367(19): 1814-1820. |
| [6] | Su S, Wong G, Shi WF, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses [J]. Trends Microbiol, 2016, 24(6): 490-502. |
| [7] | Saif LJ. Bovine respiratory coronavirus [J]. Vet Clin N Am Food Anim Pract, 2010, 26(2): 349-364. |
| [8] | Liang YW, Wang ML, Chien CS, et al. Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV, and SARS-cov-2 infection [J]. Front Immunol, 2020, 11: 1022. |
| [9] | Heckert RA, Saif LJ, Hoblet KH, et al. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio [J]. Vet Microbiol, 1990, 22(2/3): 187-201. |
| [10] | Decaro N, Campolo M, Desario C, et al. Respiratory disease associated with bovine coronavirus infection in cattle herds in southern Italy [J]. J VET Diagn Invest, 2008, 20(1): 28-32. |
| [11] | Boileau MJ, Kapil S. Bovine coronavirus associated syndromes [J]. Vet Clin N Am Food Anim Pract, 2010, 26(1): 123-146. |
| [12] | Fulton RW, Herd HR, Sorensen NJ, et al. Enteric disease in postweaned beef calves associated with Bovine coronavirus clade 2 [J]. J VET Diagn Invest, 2015, 27(1): 97-101. |
| [13] | Alekseev KP, Vlasova AN, Jung K, et al. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences [J]. J Virol, 2008, 82(24): 12422-12431. |
| [14] | Burimuah V, Sylverken A, Owusu M, et al. Molecular-based cross-species evaluation of bovine coronavirus infection in cattle, sheep and goats in Ghana [J]. BMC Vet Res, 2020, 16(1): 405. |
| [15] | Mao L, Cai XH, Li JZ, et al. Discovery of a novel Betacoronavirus 1, cpCoV, in goats in China: The new risk of cross-species transmission [J]. PLoS Pathog, 2025, 21(3): e1012974. |
| [16] | Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity [J]. Nature, 1975, 256(5517): 495-497. |
| [17] | Amer HM. Bovine-like coronaviruses in domestic and wild ruminants [J]. Anim Health Res Rev, 2018, 19(2): 113-124. |
| [18] | Saif LJ, Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans [J]. J Clin Microbiol, 2020, 58(8): e01355-20. |
| [19] | 靳双媛, 杜家伟, 王雪妍, 等. 牛冠状病毒S和HE蛋白研究进展 [J]. 中国畜牧兽医, 2024, 51(7): 3118-3127. |
| Jin SY, Du JW, Wang XY, et al. Research progress on S and HE proteins of bovine coronaviruses [J]. China Anim Husb Vet Med, 2024, 51(7): 3118-3127. | |
| [20] | Zhu QH, Li B, Sun DB. Advances in bovine coronavirus epidemiology [J]. Viruses, 2022, 14(5): 1109. |
| [21] | 刘筱筱, 余秋蓉, 李霞, 等. 牛冠状病毒家兔源多克隆抗体的制备与应用 [J/OL]. 中国兽医科学, 2025: 1-9. (2025-05-29). . |
| Liu XX, Yu QR, Li X, et al. Preparation and application of rabbit polyclonal antibodies against bovine coronavirus [J/OL]. Chin Vet Sci, 2025: 1-9. (2025-05-29). . | |
| [22] | Dong SJ, Sun JC, Mao Z, et al. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV) [J]. J Med Virol, 2020, 92(9): 1542-1548. |
| [23] | Li F. Structure, function, and evolution of coronavirus spike proteins [J]. Annu Rev Virol, 2016, 3: 237-261. |
| [24] | 黎梦帆, 李青阳, 宋艳雯, 等. 冠状病毒S蛋白的结构和功能研究进展 [J]. 畜牧兽医学报, 2025, 56(9): 4241-4252. |
| Li MF, Li QY, Song YW, et al. Structure and function of coronavirus S proteins [J]. Acta Vet Zootechnica Sin, 2025, 56(9): 4241-4252. | |
| [25] | Jackson CB, Farzan M, Chen B, et al. Mechanisms of SARS-CoV-2 entry into cells [J]. Nat Rev Mol Cell Biol, 2022, 23(1): 3-20. |
| [26] | Zhu QH, Su MJ, Li ZJ, et al. Epidemiological survey and genetic diversity of bovine coronavirus in Northeast China [J]. Virus Res, 2022, 308: 198632. |
| [27] | Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein [J]. Cell, 2020, 181(2): 281-292.e6. |
| [28] | Jung J, Bong JH, Kim TH, et al. Isolation of antibodies against the spike protein of SARS-CoV from pig serum for competitive immunoassay [J]. BioChip J, 2021, 15(4): 396-405. |
| [29] | Millet JK, Jaimes JA, Whittaker GR. Molecular diversity of coronavirus host cell entry receptors [J]. FEMS Microbiol Rev, 2021, 45(3): fuaa057. |
| [30] | Hulswit RJG, de Haan CAM, Bosch BJ. Coronavirus spike protein and tropism changes [M]//Coronaviruses. Amsterdam: Elsevier, 2016: 29-57. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | SU Xiu-min, HAN Wen-qing, WANG Jiao, LI Peng, WANG Qiu-lan, LI Wan-xing, CAO Jin-jun. Isolation, Identification, Biological Characteristics and Biocontrol Effects of Trichoderma harzianum M408 against Tomato Early Blight [J]. Biotechnology Bulletin, 2025, 41(9): 277-288. |
| [3] | LIAN Shao-jie, TANG Sheng-shuo, KANG Chuan-li, LIU Lei, ZHENG De-qiang, DU Shuai, TANG Li-wei, ZHANG Mei-xia, LIU Qiang. Isolation, Identification, Optimization of Fermentation Conditions of High-yield Tremella fuciformis Polysaccharides Enzyme-producing Strain and Its Enzyme Characteristics Analysis [J]. Biotechnology Bulletin, 2025, 41(9): 302-313. |
| [4] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [5] | ZHANG Ru, LI Yi-ming, ZHANG Tong-xi, SUN Zhan-bin, REN Qing, PAN Han-xu. Isolation and Identification of a High-yielding Magnolol and Honokiol Strain from Magnolia officinalis and Optimization of the “Sweating” Process [J]. Biotechnology Bulletin, 2025, 41(8): 322-334. |
| [6] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [7] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [8] | WANG Yi-min, LI Ying, DONG Hai-tao, ZHANG Heng-rui, CHANG Lu, GAO Tian-tian, HAN De-jun, WU Jian-hui. Evolutionary Patterns of SRO Family Proteins in the Polyploidization Process of Wheat [J]. Biotechnology Bulletin, 2025, 41(5): 70-81. |
| [9] | LI Wen-lan, HOU Xin-wei, LI Yan, ZHAO Rui-jun, MENG Zhao-dong, YUE Run-qing. Identification and Resistance Detection of Homozygous and Heterozygous Plants of Transgenic Maize LD05 with Resistances to Insect and Herbicide [J]. Biotechnology Bulletin, 2025, 41(4): 123-133. |
| [10] | SUN Tian-guo, YI Lan, QIN Xu-yang, QIAO Meng-xue, GU Xin-ying, HAN Yi, SHA Wei, ZHANG Mei-juan, MA Tian-yi. Genome-wide Identification of the DABB Gene Family in Brassica rapa ssp. pekinensis and Expression Analysis under Saline and Alkali Stress [J]. Biotechnology Bulletin, 2025, 41(4): 156-165. |
| [11] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [12] | HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties [J]. Biotechnology Bulletin, 2025, 41(4): 243-255. |
| [13] | LIU Shuang, JIANG Zhou, ZHAO Shuai, ZHAO Lei-zhen, HUANG Feng, ZHOU Jia, QU Jian-hang. Screening, Identification, and Fermentation Optimization of a Protease-producing Bacterial Strain [J]. Biotechnology Bulletin, 2025, 41(4): 335-344. |
| [14] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [15] | MA Tian-yi, XU Jia-jia, LU Wen-jing, WU Yan, SHA Wei, ZHANG Mei-juan, PENG Yi-fang. Expression Analysis and Resistance Identification of BrcGASA3 in Chinese Cabbage ‘Jinxiaotong’ Cultivar under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 127-138. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||