








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (2): 1-11.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0908
NONG Wei-you1( ), ZHAO Chang-zu1, QIAN Zhen-feng1, DING Qian1, WANG Yu-jie1, CHEN Shu-ying1,2, HE Li-lian1,2(
), ZHAO Chang-zu1, QIAN Zhen-feng1, DING Qian1, WANG Yu-jie1, CHEN Shu-ying1,2, HE Li-lian1,2( ), LI Fu-sheng1,2,3(
), LI Fu-sheng1,2,3( )
)
Received:2025-08-23
Online:2026-02-09
Published:2026-02-09
Contact:
HE Li-lian, LI Fu-sheng
E-mail:2254104913@qq.com;helilian905@sohu.com;Lfs810@sina.com
NONG Wei-you, ZHAO Chang-zu, QIAN Zhen-feng, DING Qian, WANG Yu-jie, CHEN Shu-ying, HE Li-lian, LI Fu-sheng. Identification of the EfBBX Gene Family in Erianthus fulvus and Analysis of Its Expression Patterns Under Cold Stress[J]. Biotechnology Bulletin, 2026, 42(2): 1-11.
| 基因 Gene | 基因IDGene ID | 氨基酸数量 Number of amino Acids (aa) | 相对分子量 Molecular Weight (Da) | 等电点PI | 不稳定指数 Instability index | 脂肪指数 Aliphatic index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| EfBBX1 | Erufi.01G0010670.t1 | 420 | 45 969.84 | 5.88 | 55.68 | 64.21 | 叶绿体 Chloroplast |
| EfBBX2 | Erufi.03G0003170.t1 | 358 | 37 106.31 | 5.42 | 51.18 | 65.67 | 细胞核 Nuclear |
| EfBBX3 | Erufi.04G0006700.t1 | 373 | 38 846.33 | 5.80 | 45.40 | 65.39 | 叶绿体 Chloroplast |
| EfBBX4 | Erufi.04G0019680.t1 | 257 | 27 606.01 | 4.95 | 43.19 | 76.54 | 细胞质 Cytoplasmic |
| EfBBX5 | Erufi.04G0020000.t1 | 328 | 34 599.45 | 5.12 | 44.90 | 66.10 | 细胞质 Cytoplasmic |
| EfBBX6 | Erufi.04G0022180.t1 | 276 | 28 816.93 | 5.38 | 61.93 | 63.91 | 细胞核 Nuclear |
| EfBBX7 | Erufi.04G0026320.t1 | 405 | 44 398.46 | 5.30 | 50.12 | 61.73 | 细胞核 Nuclear |
| EfBBX8 | Erufi.04G0026680.t1 | 374 | 41 614.82 | 6.42 | 56.63 | 70.16 | 叶绿体 Chloroplast |
| EfBBX9 | Erufi.04G0026930.t1 | 476 | 51 626.87 | 5.68 | 65.17 | 63.87 | 叶绿体 Chloroplast |
| EfBBX10 | Erufi.06G0013060.t1 | 255 | 27 181.54 | 4.80 | 49.61 | 69.76 | 细胞核 Nuclear |
| EfBBX11 | Erufi.06G0013390.t1 | 354 | 37 454.85 | 5.46 | 38.07 | 63.73 | 叶绿体 Chloroplast |
| EfBBX12 | Erufi.06G0016230.t1 | 261 | 28 309.31 | 4.89 | 70.95 | 61.07 | 细胞核 Nuclear |
| EfBBX13 | Erufi.09G0008130.t1 | 327 | 34 135.01 | 5.07 | 56.04 | 63.15 | 细胞核 Nuclear |
| EfBBX14 | Erufi.10G0004850.t1 | 481 | 52 067.46 | 6.60 | 61.19 | 67.53 | 叶绿体 Chloroplast |
| EfBBX15 | Erufi.10G0011520.t1 | 459 | 49 201.79 | 5.42 | 46.14 | 68.82 | 细胞质 Cytoplasmic |
| EfBBX16 | Erufi.10G0012110.t1 | 403 | 43 490.53 | 5.31 | 48.03 | 69.58 | 细胞质 Cytoplasmic |
| EfBBX17 | Erufi.10G0012650.t1 | 404 | 43 489.41 | 5.30 | 46.75 | 65.50 | 细胞核 Nuclear |
| EfBBX18 | Erufi.10G0020560.t1 | 366 | 39 089.78 | 5.76 | 43.58 | 69.48 | 细胞质 Cytoplasmic |
| EfBBX19 | Erufi.10G0025500.t1 | 303 | 31 346.08 | 4.83 | 57.21 | 70.99 | 叶绿体 Chloroplast |
Table 1 Physical and chemical properties of EfBBX gene family in Erianthusfulvus
| 基因 Gene | 基因IDGene ID | 氨基酸数量 Number of amino Acids (aa) | 相对分子量 Molecular Weight (Da) | 等电点PI | 不稳定指数 Instability index | 脂肪指数 Aliphatic index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| EfBBX1 | Erufi.01G0010670.t1 | 420 | 45 969.84 | 5.88 | 55.68 | 64.21 | 叶绿体 Chloroplast |
| EfBBX2 | Erufi.03G0003170.t1 | 358 | 37 106.31 | 5.42 | 51.18 | 65.67 | 细胞核 Nuclear |
| EfBBX3 | Erufi.04G0006700.t1 | 373 | 38 846.33 | 5.80 | 45.40 | 65.39 | 叶绿体 Chloroplast |
| EfBBX4 | Erufi.04G0019680.t1 | 257 | 27 606.01 | 4.95 | 43.19 | 76.54 | 细胞质 Cytoplasmic |
| EfBBX5 | Erufi.04G0020000.t1 | 328 | 34 599.45 | 5.12 | 44.90 | 66.10 | 细胞质 Cytoplasmic |
| EfBBX6 | Erufi.04G0022180.t1 | 276 | 28 816.93 | 5.38 | 61.93 | 63.91 | 细胞核 Nuclear |
| EfBBX7 | Erufi.04G0026320.t1 | 405 | 44 398.46 | 5.30 | 50.12 | 61.73 | 细胞核 Nuclear |
| EfBBX8 | Erufi.04G0026680.t1 | 374 | 41 614.82 | 6.42 | 56.63 | 70.16 | 叶绿体 Chloroplast |
| EfBBX9 | Erufi.04G0026930.t1 | 476 | 51 626.87 | 5.68 | 65.17 | 63.87 | 叶绿体 Chloroplast |
| EfBBX10 | Erufi.06G0013060.t1 | 255 | 27 181.54 | 4.80 | 49.61 | 69.76 | 细胞核 Nuclear |
| EfBBX11 | Erufi.06G0013390.t1 | 354 | 37 454.85 | 5.46 | 38.07 | 63.73 | 叶绿体 Chloroplast |
| EfBBX12 | Erufi.06G0016230.t1 | 261 | 28 309.31 | 4.89 | 70.95 | 61.07 | 细胞核 Nuclear |
| EfBBX13 | Erufi.09G0008130.t1 | 327 | 34 135.01 | 5.07 | 56.04 | 63.15 | 细胞核 Nuclear |
| EfBBX14 | Erufi.10G0004850.t1 | 481 | 52 067.46 | 6.60 | 61.19 | 67.53 | 叶绿体 Chloroplast |
| EfBBX15 | Erufi.10G0011520.t1 | 459 | 49 201.79 | 5.42 | 46.14 | 68.82 | 细胞质 Cytoplasmic |
| EfBBX16 | Erufi.10G0012110.t1 | 403 | 43 490.53 | 5.31 | 48.03 | 69.58 | 细胞质 Cytoplasmic |
| EfBBX17 | Erufi.10G0012650.t1 | 404 | 43 489.41 | 5.30 | 46.75 | 65.50 | 细胞核 Nuclear |
| EfBBX18 | Erufi.10G0020560.t1 | 366 | 39 089.78 | 5.76 | 43.58 | 69.48 | 细胞质 Cytoplasmic |
| EfBBX19 | Erufi.10G0025500.t1 | 303 | 31 346.08 | 4.83 | 57.21 | 70.99 | 叶绿体 Chloroplast |
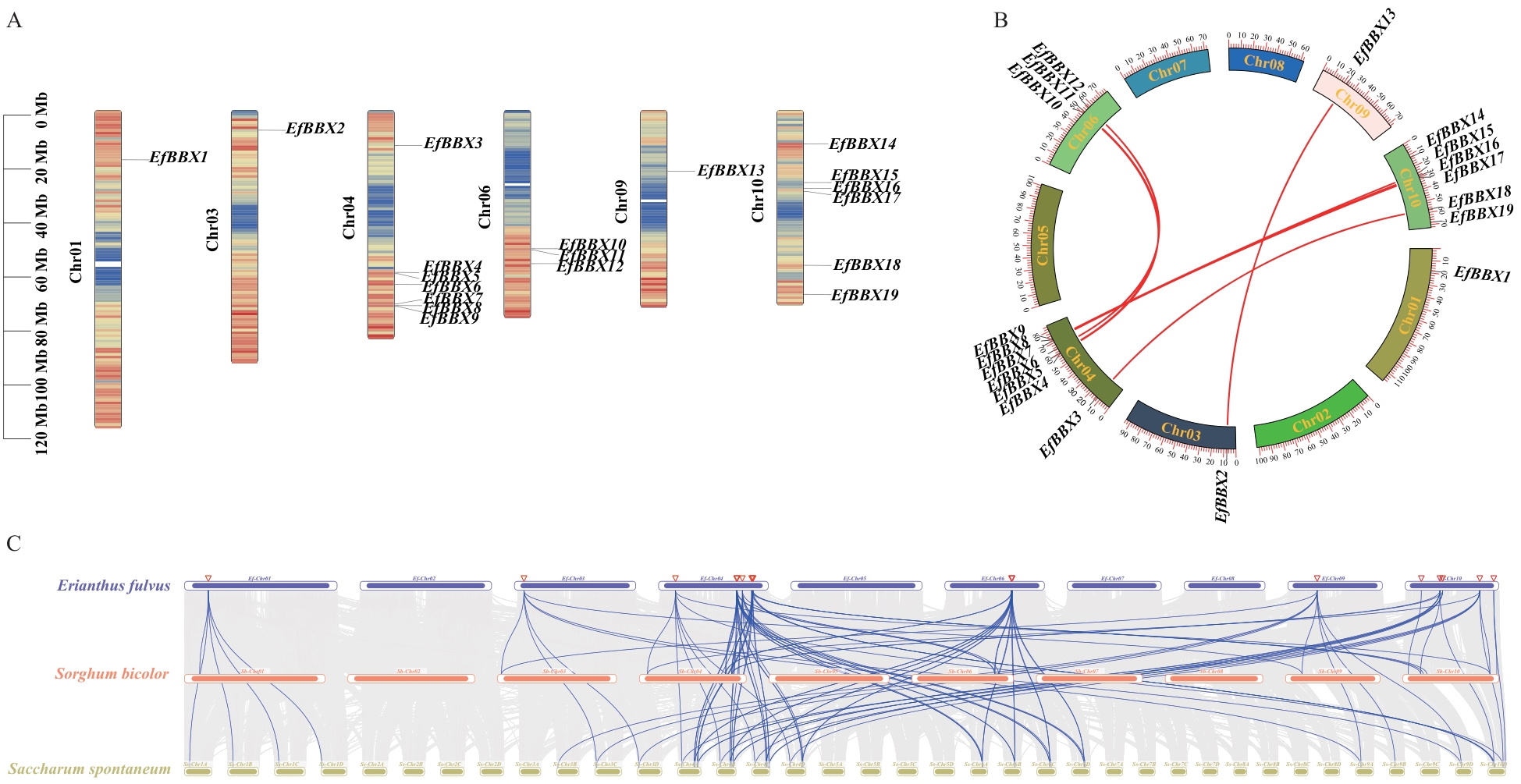
Fig. 2 Chromosomal localization and colinearity analysis of the EfBBX Gene in E.fulvusA: Mapping of EfBBX gene on chromosomes. B: Intraspecific collinearity analysis of E.fulvusEfBBX genes. C: Collinearity analysis among E.fulvus, Sorghum bicolor, and Saccharum spontaneum

Fig. 7 E. fulvus's EfBBX18 transiently expressed in Nicotiana benthamianaA: Tobacco plants after 0 h and 24 h of low-temperature stress treatment. B: RT-qPCR results
| [1] | 高松, 王鹤潼. 低温胁迫下植物调控转录因子家族研究进展 [J]. 世界热带农业信息, 2025(6): 111-114. |
| Gao S, Wang HT. Research progress of plant regulation transcription factor family under low temperature stress [J]. World Trop Agric Inf, 2025(6): 111-114. | |
| [2] | 郭佳强, 曾鑫, 陈孝平. 植物抗低温胁迫的研究进展 [J]. 武汉工程大学学报, 2023, 45(2): 119-125. |
| Guojia Q, Zeng X, Chen XP. Advances in plant resistance to low temperature stress [J]. J Wuhan Inst Technol, 2023, 45(2): 119-125. | |
| [3] | 李子涵, 李建运, 王飞, 等. 植物对低温胁迫反应机制的研究进展 [J]. 农业与技术, 2024, 44(7): 134-138. |
| Li ZH, Li JY, Wang F, et al. Research progress on response mechanism of plants to low temperature stress [J]. Agric Technol, 2024, 44(7): 134-138. | |
| [4] | 赵婷婷, 孙婷婷, 王俊刚, 等. 甘蔗育种研究进展 [J]. 中国科学: 生命科学, 2024, 54(10): 1814-1832. |
| Zhao TT, Sun TT, Wang JG, et al. The research progress on sugarcane breeding [J]. Sci Sin Vitae, 2024, 54(10): 1814-1832. | |
| [5] | 周慧文, 陆桂军, 吴建明, 等. 中国甘蔗种业发展研究进展 [J]. 广西科学, 2023, 30(3): 421-433. |
| Zhou HW, Lu GJ, Wu JM, et al. Research progress of sugarcane seed industry in China [J]. Guangxi Sci, 2023, 30(3): 421-433. | |
| [6] | 胡水凤. 甘蔗抗逆转基因研究进展 [J]. 广西糖业, 2022, 42(5): 18-21. |
| Hu SF. Research progress of sugarcane anti-reversion genes [J]. Guangxi Sugar Ind, 2022, 42(5): 18-21. | |
| [7] | 张志勇, 雷朝云, 蒙秋伊, 等. 甘蔗抗逆基因工程育种研究进展 [J]. 核农学报, 2012, 26(3): 471-477. |
| Zhang ZY, Lei CY, Meng QY, et al. Research progress on genetic engineering of stress tolerance in sugarcane [J]. J Nucl Agric Sci, 2012, 26(3): 471-477. | |
| [8] | 萧凤回, 李富生, 何丽莲, 等. 甘蔗近缘野生种蔗茅(Erianthus rufipilus)的研究 [J]. 甘蔗, 1996(2): 1-6. |
| Xiao FH, Li FS, He LL, et al. Study on the wild species of sugarcane (Erianthus rufipilus) [J]. Subtrop Agric Res, 1996(2): 1-6. | |
| [9] | 李富生, 林位夫, 何顺长. 开发利用蔗茅野生种质资源的思考 [J]. 资源开发与市场, 2004, 20(4): 266-270. |
| Li FS, Lin WF, He SC. Suggestions on developing and utilization of Erianthus fulvus wild species [J]. Resour Dev Mark, 2004, 20(4): 266-270. | |
| [10] | 王丽萍, 蔡青, 陆鑫, 等. 甘蔗近缘属野生种滇蔗茅(Erianthus rockii)的种质创新利用 [J]. 中国糖料, 2008, 30(2): 8-11. |
| Wang LP, Cai Q, Lu X, et al. Study of wild species Erianthus rockii germplasm innovation and use [J]. Sugar Crops China, 2008, 30(2): 8-11. | |
| [11] | 王秋松. 甘蔗与滇蔗茅后代染色体遗传及利用价值研究 [D]. 福州: 福建农林大学, 2022. |
| Wang QS. Research on chromosome inheritance and application value of the progenies of Saccharum officinarum and Erianthus rockii [D]. Fuzhou: Fujian Agriculture and Forestry University, 2022. | |
| [12] | 钱禛锋, 赵昌祖, 万慧兰, 等. 甘蔗近缘野生种蔗茅的研究进展和利用潜力 [J]. 植物遗传资源学报, 2025, 26(8): 1485-1498. |
| Qian ZF, Zhao CZ, Wan HL, et al. Research progress and utilization potential of wild sugarcane species Erianthus fulvus ness [J]. J Plant Genet Resour, 2025, 26(8): 1485-1498. | |
| [13] | 杨宁, 从青, 程龙军. 植物BBX转录因子基因家族的研究进展 [J]. 生物工程学报, 2020, 36(4): 666-677. |
| Yang N, Cong Q, Cheng LJ. BBX transcriptional factors family in plants-a review [J]. Chin J Biotechnol, 2020, 36(4): 666-677. | |
| [14] | Gangappa SN, Botto JF. The BBX family of plant transcription factors [J]. Trends Plant Sci, 2014, 19(7): 460-470. |
| [15] | Gao LL, Xu S, Zhang JM, et al. Promotion of seedling germination in Arabidopsis by B-box zinc-finger protein BBX32 [J]. Curr Biol, 2024, 34(14): 3152-3164.e6. |
| [16] | Song ZQ, Bian YT, Liu JJ, et al. B-box proteins: Pivotal players in light-mediated development in plants [J]. J Integr Plant Biol, 2020, 62(9): 1293-1309. |
| [17] | Cao J, Liang YX, Yan TT, et al. The photomorphogenic repressors BBX28 and BBX29 integrate light and brassinosteroid signaling to inhibit seedling development in Arabidopsis [J]. Plant Cell, 2022, 34(6): 2266-2285. |
| [18] | Bian YT, Song ZL, Liu CS, et al. The BBX7/8-CCA1/LHY transcription factor cascade promotes shade avoidance by activating PIF4 [J]. New Phytol, 2025, 245(2): 637-652. |
| [19] | Song ZQ, Yan TT, Liu JJ, et al. BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development [J]. Plant J, 2020, 104(2): 377-390. |
| [20] | Yang CL, Zhang SW, Zou QJ, et al. CiBBX19 negatively regulates the flowering time of Chrysanthemum indicum by recruiting CiBBX5 to inhibit the expression of CiFTL3 [J]. Plant Physiol Biochem, 2025, 227: 110115. |
| [21] | Fu JL, Liao L, Jin JJ, et al. A transcriptional cascade involving BBX22 and HY5 finely regulates both plant height and fruit pigmentation in Citrus [J]. J Integr Plant Biol, 2024, 66(8): 1752-1768. |
| [22] | 邓军, 罗正良, 朱菲莹, 等. 水稻B-box锌指蛋白基因OsBBX26的克隆及表达分析 [J]. 南方农业学报, 2020, 51(7): 1606-1613. |
| Deng J, Luo ZL, Zhu FY, et al. Cloning and expression analysis of B-box zinc-finger protein gene OsBBX26 in rice [J]. J South Agric, 2020, 51(7): 1606-1613. | |
| [23] | 齐小坤, 孟祥熹, 赵志明, 等. 毛果杨BBX基因家族鉴定与生物信息学分析 [J]. 河北农业大学学报, 2023, 46(5): 41-49. |
| Qi XK, Meng XX, Zhao ZM, et al. Identification and bioinformatics analysis of BBX gene family in Populus trichocarpa [J]. J Hebei Agric Univ, 2023, 46(5): 41-49. | |
| [24] | 宁露云, 李蓓, 李佩, 等. 矮牵牛B-box型锌指蛋白基因PhBBX8的克隆与表达分析 [J]. 园艺学报, 2014, 41(12): 2437-2445. |
| Ning LY, Li B, Li P, et al. Cloning and expression analysis of a B-box-type zinc-finger protein gene PhBBX8 in Petunia hybrida [J]. Acta Hortic Sin, 2014, 41(12): 2437-2445. | |
| [25] | Li YP, Shi YT, Li MZ, et al. The CRY2-COP1-HY5-BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis [J]. Plant Cell, 2021, 33(11): 3555-3573. |
| [26] | An JP, Wang XF, Zhang XW, et al. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation [J]. New Phytol, 2021, 229(5): 2707-2729. |
| [27] | Zhang H, Chen MJ, Luo XB, et al. Overexpression of StBBX14 enhances cold tolerance in potato [J]. Plants, 2024, 14(1): 18. |
| [28] | Song JN, Lin R, Tang MJ, et al. SlMPK1- and SlMPK2-mediated SlBBX17 phosphorylation positively regulates CBF-dependent cold tolerance in tomato [J]. New Phytol, 2023, 239(5): 1887-1902. |
| [29] | Bu X, Wang XJ, Yan JR, et al. Genome-wide characterization of B-box gene family and its roles in responses to light quality and cold stress in tomato [J]. Front Plant Sci, 2021, 12: 698525. |
| [30] | Zhu YF, Zhu GT, Xu R, et al. A natural promoter variation of SlBBX31 confers enhanced cold tolerance during tomato domestication [J]. Plant Biotechnol J, 2023, 21(5): 1033-1043. |
| [31] | Wang S, Shen YR, Deng DY, et al. Orthogroup and phylotranscriptomic analyses identify transcription factors involved in the plant cold response: a case study of Arabidopsis BBX29 [J]. Plant Commun, 2023, 4(6): 100684. |
| [32] | Lyu GZ, Li DB, Li SS. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana [J]. Plant Signal Behav, 2020, 15(9): 1782647. |
| [33] | Huang JY, Zhao XB, Weng XY, et al. The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis [J]. PLoS One, 2012, 7(10): e48242. |
| [34] | Shalmani A, Jing XQ, Shi Y, et al. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants [J]. BMC Genomics, 2019, 20(1): 27. |
| [35] | Chu ZN, Wang X, Li Y, et al. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato [J]. Front Plant Sci, 2016, 7: 1552. |
| [36] | Yin LL, Wu RG, An RL, et al. Genome-wide identification, molecular evolution and expression analysis of the B-box gene family in mung bean (Vigna radiata L.) [J]. BMC Plant Biol, 2024, 24(1): 532. |
| [37] | Kui L, Majeed A, Wang XH, et al. A chromosome-level genome assembly for Erianthus fulvus provides insights into its biofuel potential and facilitates breeding for improvement of sugarcane [J]. Plant Commun, 2023, 4(4): 100562. |
| [38] | Singh S, Chhapekar SS, Ma YB, et al. Genome-wide identification, evolution, and comparative analysis of B-Box genes in Brassica rapa, B. oleracea, and B. napus and their expression profiling in B. Rapa in response to multiple hormones and abiotic stresses [J]. Int J Mol Sci, 2021, 22(19): 10367. |
| [39] | Liu Y, Wang YX, Liao J, et al. Identification and characterization of the BBX gene family in Bambusa pervariabilis × Dendrocalamopsis grandis and their potential role under adverse environmental stresses [J]. Int J Mol Sci, 2023, 24(17): 13465. |
| [40] | 马文婧, 刘震, 李志涛, 等. 马铃薯BBX基因家族的全基因组鉴定及表达分析 [J]. 作物学报, 2022(11): 2797-2812. |
| Ma WJ, Liu Z, Li ZT, et al. Whole genome identification and expression analysis of potato BBX gene family [J]. Acta Agronomica Sinica, 2022(11): 2797-2812. | |
| [41] | 殷丽丽, 邢宝龙, 陈晓亮, 等. 大豆BBX基因家族的鉴定及表达 [J]. 西北农林科技大学学报: 自然科学版, 2022, 50(6): 35-45. |
| Yin LL, Xing BL, Chen XL, et al. Genome-wide identification and expression patterns of BBX gene family members in Glycine max [J]. J Northwest A F Univ Nat Sci Ed, 2022, 50(6): 35-45. | |
| [42] | Liu X, Li R, Dai YQ, et al. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome [J]. Mol Genet Genomics, 2018, 293(2): 303-315. |
| [43] | 翟海涛, 余月, 陈蕾, 等. 芥菜BjuBBX基因家族全基因组鉴定与分析 [J]. 江西农业学报, 2023, 35(11): 40-50. |
| Zhai HT, Yu Y, Chen L, et al. Genome identification and analysis of BjuBBX gene family in mustard [J]. Acta Agric Jiangxi, 2023, 35(11): 40-50. | |
| [44] | Wu ZL, Fu DW, Gao XN, et al. Characterization and expression profiles of the B-box gene family during plant growth and under low-nitrogen stress in Saccharum [J]. BMC Genomics, 2023, 24(1): 79. |
| [45] | Cheng XR, Lei SY, Li J, et al. In silico analysis of the wheat BBX gene family and identification of candidate genes for seed dormancy and germination [J]. BMC Plant Biol, 2024, 24(1): 334. |
| [46] | 杨倩, 苏旭, 刘玉萍, 等. 沙鞭SnRK2基因家族鉴定及干旱胁迫下表达分析 [J]. 草地学报, 2025, 33(10): 3173-3184. |
| Yang Q, Su X, Liu YP, et al. Identification of SnRK2 gene family in Psammochloa villosa and analysis of their expression under drought stress [J]. Acta Agrestia Sin, 2025, 33(10): 3173-3184. | |
| [47] | Chandra Malakar B, Singh S, Garhwal V, et al. BBX24 and BBX25 antagonize the function of thermosensor ELF3 to promote PIF4-mediated thermomorphogenesis in Arabidopsis [J]. Plant Commun, 2025, 6(7): 101391. |
| [48] | Gao XR, Zhang H, Li X, et al. The B-box transcription factor IbBBX29 regulates leaf development and flavonoid biosynthesis in sweet potato [J]. Plant Physiol, 2023, 191(1): 496-514. |
| [49] | Feng XJ, Li JF, Tang ZR, et al. BjuBBX6-1 interacts with BjuNF-YB2/3 to regulate flowering time and drought tolerance in Brassica juncea [J]. Hortic Plant J, 2025 |
| [50] | Bandara WW, Wijesundera WSS, Hettiarachchi C. Rice and Arabidopsis BBX proteins: toward genetic engineering of abiotic stress resistant crops [J]. 3 Biotech, 2022, 12(8): 164. |
| [51] | Dai YQ, Lu Y, Zhou Z, et al. B-box containing protein 1 from Malus domestica (MdBBX1) is involved in the abiotic stress response [J]. PeerJ, 2022, 10: e12852. |
| [52] | Qian ZF, He LL, Li FS. Understanding cold stress response mechanisms in plants: an overview [J]. Front Plant Sci, 2024, 15: 1443317. |
| [1] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| [2] | YANG Dan, JIN Ya-rong, MAO Chun-li, WANG Bi-xian, ZHANG Ya-ning, YANG Zhi-yi, ZHOU Zhi-yao, YANG Rui-ming, FAN Heng-rui, HUANG Lin-kai, YAN Hai-dong. Identification and Expression Analysis of C2H2 Gene Family in Cenchrus purpureus [J]. Biotechnology Bulletin, 2026, 42(1): 251-261. |
| [3] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [4] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [5] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [6] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [7] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [8] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [9] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [10] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [11] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [12] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [13] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [14] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [15] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||