








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 195-202.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1361
Previous Articles Next Articles
Received:2020-11-05
Online:2021-08-26
Published:2021-09-10
Contact:
DING Yu
E-mail:980930967@qq.com;dingy@gdou.edu.cn
FAN Chen-long, DING Yu. Molecular Cloning and Functional Verification of Histone Deacetylase Gene cobB in Vibrio alginolyticus[J]. Biotechnology Bulletin, 2021, 37(8): 195-202.
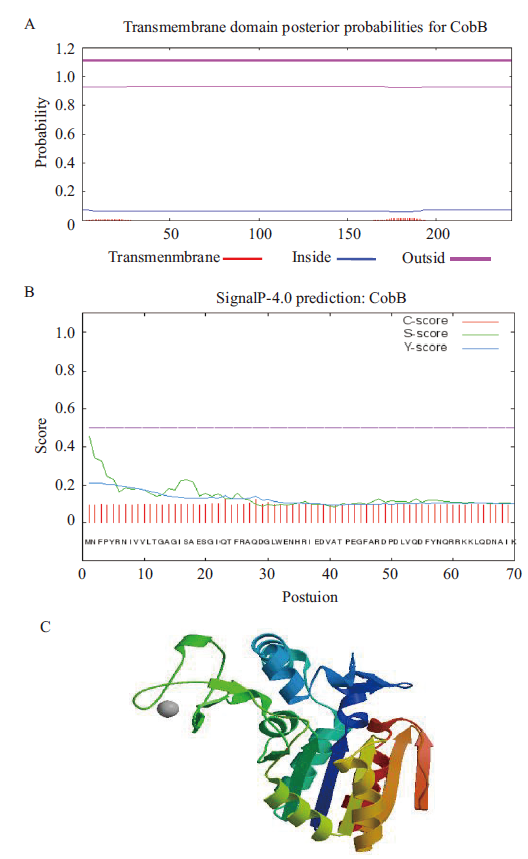
Fig.2 Bioinformatics analysis of cobB sequences A:Signal peptide prediction. B:Transmembrane domain prediction.C:Protein tertiary structure prediction
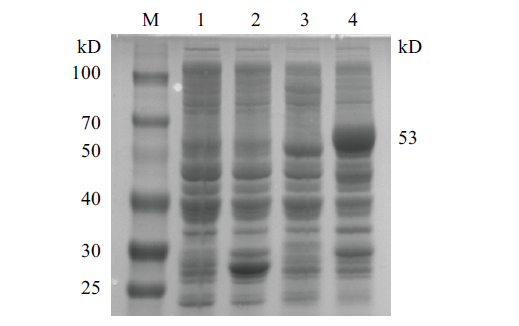
Fig.4 Schematic diagram of gel electrophoresis of CobB protein expression M:Protein marker;1:negative control without IPTG;2:negative control with IPTG;3:pGEX-6p-cobB without IPTG;4:pGEX-6p-cobB with IPTG
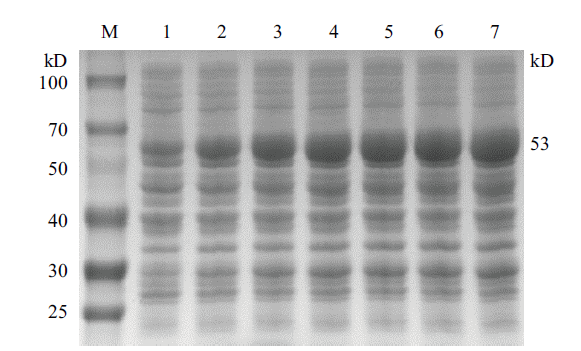
Fig.5 Schematic diagram of gel electrophoresis of different induction time on CobB protein expression M:Protein marker;1-7:The induction time is 1 h、2 h、3 h、4 h、5 h、6 h、7 h
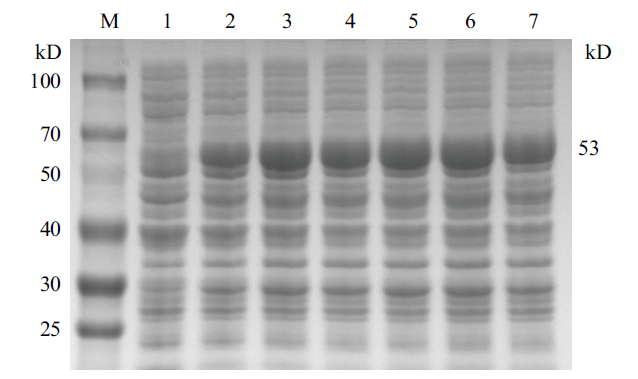
Fig.6 Schematic diagram of gel electrophoresis of different IPTG concentrations on CobB protein expression M:Protein marker;1-7:the IPTG concentrations are 0.1%、0.2%、0.4%、0.6%、0.8%、1% and 2%

Fig.7 Schematic diagram of gel electrophoresis of different temperatures on CobB protein expression M:Protein maker;1:28℃ bacteria protein;2:28℃ supernatant protein;3:28℃ precipitation protein;4:37℃ bacteria protein;5:37℃ supernatant protein;6:37℃ precipitation protein
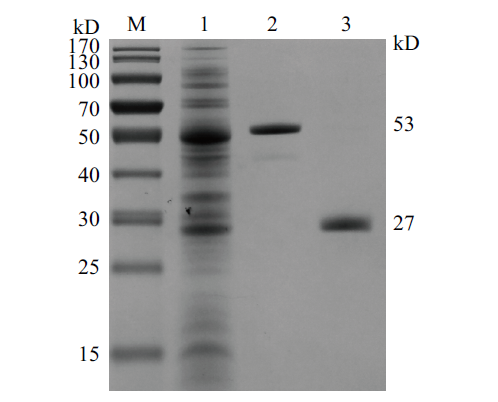
Fig.8 Schematic diagram of digestion gel electrophoresis of CobB protein purification M:Protein molecular standard;1:Bacteria protein;2:Results of CobB fusion protein purification;3:Results of CobB fusion protease cleavage results
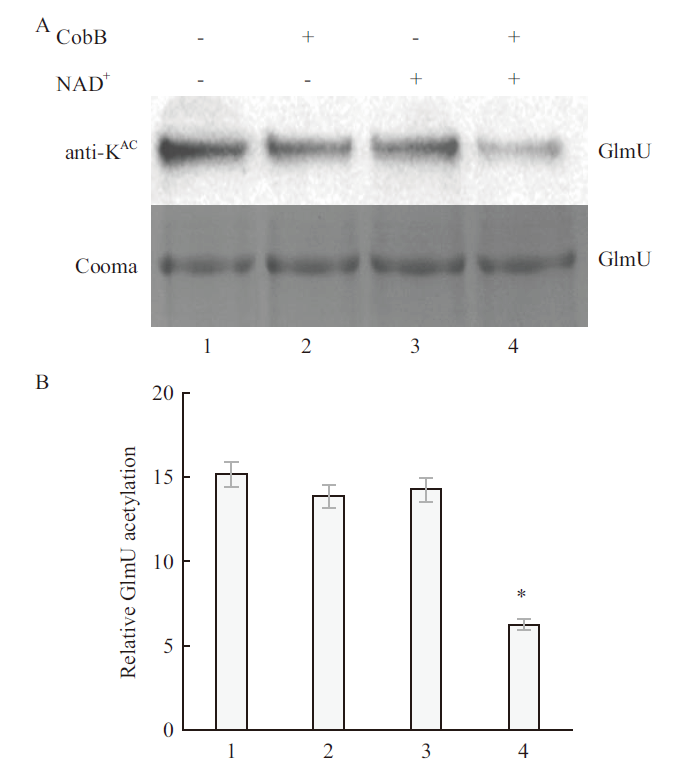
Fig.9 Western blot of CobB deacetylated GlmU protein A:Western blot of acetyllysine-specific antibodies and SDS-PAGE;B:GlmU acetylation level quantified by Western blot. 1:No CobB protein,no NAD+ group;2:CobB protein,no NAD + group;3:no CobB protein,NAD+ group;4:CobB protein,NAD+ group
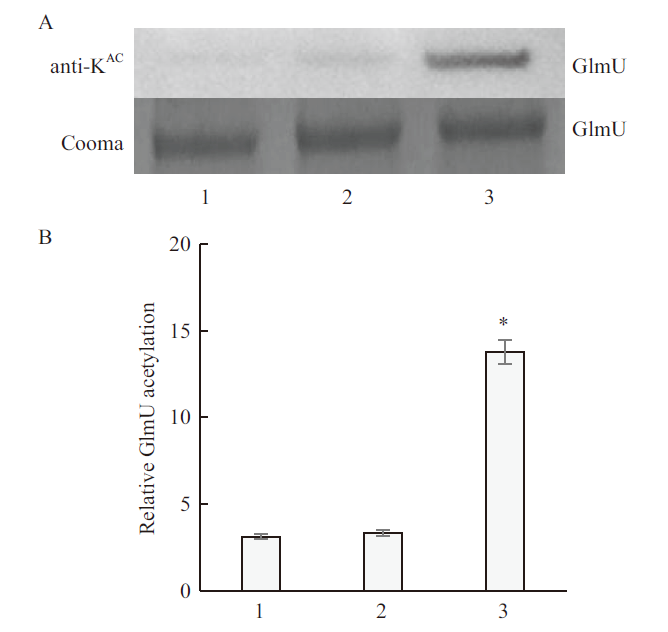
Fig.10 Western blot of CobB fusion protein and CobB protein deacetylated GlmU protein A:Western blot of acetyllysine-specific antibodies and SDS-PAGE;B:GlmU acetylation level quantified by Western blot. 1:CobB fusion protein group;2:CobB protein group;3:GST protein group
| [1] | 常云胜. 溶藻弧菌Ⅲ型分泌系统vscB基因的功能研究及vscD基因的克隆表达[D]. 湛江:广东海洋大学, 2018. |
| Chang YS. Cloning and expression of vscD gene and study on the function of vscB gene in type III secretion system of Vibrio alginolyticus[D]. Zhanjiang:Guangdong Ocean University, 2018. | |
| [2] |
Minguez P, Parcaet L, Diella F, et al. Deciphering a global network of functionally associated post-translational modifications[J]. Molecular Systems Biology, 2012, 8:599.
doi: 10.1038/msb.2012.31 pmid: 22806145 |
| [3] |
Ramponi G, Manao G, Camici G, et al. Nonenzymic acetylation of histones with acetyl phosphate and acetyl adenylate[J]. Biochemistry, 1975, 14(12):2681-2685.
pmid: 238571 |
| [4] |
Zhang L, Yao WL, Xia J, et al. Glucagon-induced acetylation of energy-sensing factors in control of hepatic metabolism[J]. International Journal of Molecular Sciences, 2019, 20:8.
doi: 10.3390/ijms20010008 URL |
| [5] |
Santos JD, Zuma AA, Vitorino FND, et al. Trichostatin A induces Trypanosoma cruzi histone and tubulin acetylation:effects on cell division and microtubule cytoskeleton remodelling[J]. Parasitology, 2019, 146(4):543-552.
doi: 10.1017/S0031182018001828 URL |
| [6] | Zhao GY, Wang H, Xu CZ, et al. SIRT6 delays cellular senescence by promoting p27Kip1 ubiquitin-proteasome degradation[J]. Aging-US, 2016, 8(10):2308-2323. |
| [7] |
Chuang C, Lin SH, Huang F, et al. Acetylation of RNA processing proteins and cell cycle proteins in mitosis[J]. Journal of Proteome Research, 2010, 9(9):4554-4564.
doi: 10.1021/pr100281h pmid: 20812760 |
| [8] |
Jiang W, Jiang P, Yang R, et al. Functional role of SIRT1-induced HMGB1 expression and acetylation in migration, invasion and angiogenesis of ovarian cancer[J]. European Review for Medical and Pharmacological Sciences, 2018, 22(14):4431-4439.
doi: 15494 pmid: 30058682 |
| [9] |
Ellinger J, Schneider AC, Bachmann A, et al. Evaluation of global histone acetylation levels in bladder cancer patients[J]. Anticancer Research, 2016, 36(8):3961-3964.
pmid: 27466500 |
| [10] |
Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins[J]. Biochemical and Biophysical Research Communications, 2000, 273(2):793-798.
pmid: 10873683 |
| [11] |
Tsang AW, Escalante-Semerena JC. CobB, a new member of the sir2 family of eucaryotic regulatory proteins, Is required to compensate for the lack of nicotinate mononucleotide:5, 6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynjournal in Salmonella typhimurium LT2[J]. Journal of Biological Chemistry, 1998, 273(48):31788-31794.
pmid: 9822644 |
| [12] |
Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene:Sir2- like proteins(sirtuins)metabolize NAD and may have protein ADP-ribosyltransferase activity[J]. Biochemical and Biophysical Research Communications, 1999, 260(1):273-279.
pmid: 10381378 |
| [13] |
Tanny JC, Dowd GJ, Huang J, et al. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing[J]. Cell, 1999, 99(7):735-745.
pmid: 10619427 |
| [14] |
Zhao KH, Chai XM, Marmorstein R, et al. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli[J]. Journal of Molecular Biology, 2004, 337(3):731-741.
doi: 10.1016/j.jmb.2004.01.060 URL |
| [15] | Gardner JG, Escalante-Semerena JC. In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene(formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase[J]. Journal of Acteriology, 2009, 191(6):1749-1755. |
| [16] |
Crosby HA, Heiniger EK, Harwood CS, et al. Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris[J]. Molecular Microbiology, 2010, 76(4):874-888.
doi: 10.1111/j.1365-2958.2010.07127.x pmid: 20345662 |
| [17] |
Karel M, Jurgen F, Eva K, et al. CobB1 deacetylase activity in Streptomyces coelicolor[J]. Biochemistry and Cell Biology, 2012, 90(2):179-187.
doi: 10.1139/o11-086 URL |
| [18] |
Starai VJ, Celic I, Cole RN, et al. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine[J]. Science, 2002, 298(5602):2390-2392.
pmid: 12493915 |
| [19] |
Kremer M, Kuhlmann N, Lechner M, et al. Comment on ‘YcgC represents a new protein deacetylase family in prokaryotes’[J]. eLife, 2018, 7:e37798.
doi: 10.7554/eLife.37798 URL |
| [20] |
AbouElfetouh A, Kuhn ML, Hu LI, et al. The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites[J]. Microbiology Open, 2015, 4(1):66-83.
doi: 10.1002/mbo3.2015.4.issue-1 URL |
| [21] | 谷晶. 2013年湖北省暨武汉微生物学会会员代表大会暨学术年会论文摘要集[C]. 武汉: 2013. |
| Gu J. 2013 Hubei Province and Wuhan society of microbiology member representative conference and academic annual conference abstracts[C]. Wuhan: 2013. | |
| [22] | 张群芳. 赖氨酸乙酰化调节大肠杆菌NhoA蛋白活性的研究[D]. 武汉:华中农业大学, 2013. |
| Zhang QF. Reversible lysine acetylation regulates the activity of E. coli N-hydroxyarylamine O-acetyltransferase[D]. Wuhan:Huazhong Agricultural University, 2013. | |
| [23] | 孙曼銮. 乙酰化调节大肠杆菌S-腺苷甲硫氨酸合成酶活性的机制研究[D]. 中国科学院研究生院, 2016. |
| Sun ML. Acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase[D]. Wuhan:University of Chinese Academy of Sciences, 2016. | |
| [24] |
Sun ML, Guo HS, Lu GL, et al. Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase[J]. Acta Biochimica et Biophysica Sinica, 2016, 48(8):723-731.
doi: 10.1093/abbs/gmw066 URL |
| [25] |
Zhou QX, Zhou YN, Jin DJ, et al. Deacetylation of topoisomerase I is an important physiological function of E-coli CobB[J]. Nucleic Acids Research, 2017, 45(9):5349-5358.
doi: 10.1093/nar/gkx250 URL |
| [26] | Xu ZW, Zhang HN, Zhang XR, et al. Interplay between the bacterial protein deacetylase CobB and the second messenger c-di-GMP[J]. EMBO Journal, 2019, 38:18. |
| [27] |
Venkat S, Gregory C, Gan QL, et al. Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli[J]. Chembiochem, 2017, 18(19):1928-1934.
doi: 10.1002/cbic.v18.19 URL |
| [28] | Venkat S, Chen H, McGuire P, et al. Characterizing lysine acetylation of Escherichia coli type II citrate synthase[J]. FEBS J, 2019, 286(14):2799-2808. |
| [29] | Ren J, Sang Y, Lu J, et al. Protein acetylation and its role in bacterial virulence[J]. Trends Microbiol, 2017, 9, 25(9):768-779. |
| [1] | YANG Jia-hui, SUN Yu-ping, LU Ya-ning, LIU huan, LU Cun-fu, CHEN Yu-zhen. Abiotic Stress Resistance of Escherichia coli Transformed with Arabidopsis thaliana AtTERT Gene [J]. Biotechnology Bulletin, 2022, 38(2): 1-9. |
| [2] | ZENG Fu-yuan, SU Ze-hui, ZHOU Shi-hui, XIE Miao, PANG Huan-ying. Prokaryotic Expression of the PEPCK Protein of Vibrio alginolyticus and Identification of Its Acetylation and Succinylation [J]. Biotechnology Bulletin, 2021, 37(5): 84-91. |
| [3] | DAI Wen-shuang, LIU Hui-yun, DU Qing-guo, ZOU Cheng, WANG Ke. Effect of Histone Deacetylase Inhibitor(HDACi)on CRISPR Editing Efficiency of Wheat and Transcriptomics Analysis [J]. Biotechnology Bulletin, 2021, 37(1): 2-14. |
| [4] | YANG Shi-quan, PENG Dan, FEI Wen-jie, YANG Feng, QU Gao-yi, TANG Wei-wei, OU Jian-ping, DENG Xiang-wen, ZHOU Bo. Cloning and Expression of ClKptA/Tpt1 Gene from Cunninghamia lanceolata(Lamb.)Hook [J]. Biotechnology Bulletin, 2020, 36(8): 15-22. |
| [5] | MIN Qi, GAO Zi-han, YAO Yin, ZHANG Hua-shan, XIONG Hai-rong, ZHANG Li. Effect of Co-expression of HAC1 and Molecular Chaperone Genes on the Expression of Mannanase in Pichia pastoris [J]. Biotechnology Bulletin, 2020, 36(5): 159-168. |
| [6] | XU Zhou, FAN Chen-long, DING Yu. Construction of Prokaryotic Expression Vector of PepA Protein of Vibrio alginolyticus and Identification of Its Acetylation [J]. Biotechnology Bulletin, 2020, 36(12): 75-81. |
| [7] | ZHONG Li-ting, CHEN Xiu-zhen, TANG Yun, LI Jun-ren, WANG Xiao-bing, LIU Yan-ting, ZHOU Xuan-xuan, ZHAN Ruo-ting, CHEN Li-kai. Expression of FPPS Recombinant Protein from Pogostemon cablin and Screening of the Interaction Proteins [J]. Biotechnology Bulletin, 2019, 35(12): 10-15. |
| [8] | GAO Qing-hua, DONG Cong, WANG Yue, HU Mei-rong, WANG Qing-qing, WANG Yun-peng, LUO Tong-yang, LIU Lei. Enhancement of Glucose Oxidase in Pichia pastoris by Co-expressing Chaperone PDI and Ero1 [J]. Biotechnology Bulletin, 2018, 34(7): 174-179. |
| [9] | WAN Qing-qing, LI Jie, ZENG Yu, ZHANG Ming-ming, ZHAO Xin-qing, BAI Feng-wu. Enhanced Production of Cellobiohydrolase in the Recombinant Saccharomyces cerevisiae by MRP8 Overexpression [J]. Biotechnology Bulletin, 2018, 34(5): 94-100. |
| [10] | ZHOU Qin-mao, XIE Yan-chun, CHEN Yan-mei, ZHANG Yang, KE De-sen, CHEN Qiong-hua. Screening and Identification of an Effective Agarase-producing Marine Bacterium [J]. Biotechnology Bulletin, 2018, 34(12): 140-146. |
| [11] | LIU Yi-jun, JIA Yu-kun, WANG Ling-fang, LIU Hong-xing, YANG Xian-yu. Prokaryotic Expression,Purification and Antiserum Preparation of Recombinant EDF-1 of Bufo gargarizans [J]. Biotechnology Bulletin, 2018, 34(10): 129-134. |
| [12] | LIU Yan-xia,LI Shao-jie,FAN Zhen-chuan. Prokaryotic Expression,Purification and Polyclonal Antibody Preparation of Neurospora crassa ERG-11 Protein Antigen [J]. Biotechnology Bulletin, 2017, 33(9): 216-222. |
| [13] | LI Yu-feng, DAI Xi-lin,YUAN Xin-cheng,ZHOU Xun,HU Yan-jie,DING Fu-jiang. Comparing and Analyzing the Disease Resistances of Four Disease-resistant Selected Lines of Macrobrachium rosenbergii [J]. Biotechnology Bulletin, 2017, 33(7): 203-209. |
| [14] | ZHAO Shu-mei, WANG Lin-hui, TANG Jia-qi, ZHAO Jing-yi. Cloning,Expression and Characterization of the Esterase Gene estZ from Vibrio alginolyticus [J]. Biotechnology Bulletin, 2017, 33(6): 190-196. |
| [15] | AN Meng-nan, WU Yuan-hua. Research Progress on BYL Cell-free Systems in Plant RNA Virus Translation and Replication Mechanisms [J]. Biotechnology Bulletin, 2017, 33(12): 45-50. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
