








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 42-56.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1158
Previous Articles Next Articles
CHEN Ti-qiang1( ), XU Xiao-lan2, SHI Lin-chun3, ZHONG Li-Yi4
), XU Xiao-lan2, SHI Lin-chun3, ZHONG Li-Yi4
Received:2021-09-09
Online:2021-11-26
Published:2021-12-03
Contact:
CHEN Ti-qiang
E-mail:chen_tiqiang@189.cn
CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2)[J]. Biotechnology Bulletin, 2021, 37(11): 42-56.
| 数据Data | Read总条数 Total number of reads | 碱基总数目 Total base/bp | Reads长度 Reads length/bp | GC% | Q20* | Q30** |
|---|---|---|---|---|---|---|
| Raw data | 61 457 184 | 9 218 577 600 | 150∶150 | 54.37 | 97.49 | 93.09 |
| Clean data | 55 973 534 | 8 396 030 100 | 150∶150 | 54.36 | 97.33 | 92.81 |
Table 1 Statistics of second generation sequencing data(Illumina Nova Seq6000)and filtering results
| 数据Data | Read总条数 Total number of reads | 碱基总数目 Total base/bp | Reads长度 Reads length/bp | GC% | Q20* | Q30** |
|---|---|---|---|---|---|---|
| Raw data | 61 457 184 | 9 218 577 600 | 150∶150 | 54.37 | 97.49 | 93.09 |
| Clean data | 55 973 534 | 8 396 030 100 | 150∶150 | 54.36 | 97.33 | 92.81 |
| 项目 Item | 数据 Data | |
|---|---|---|
| 下机的碱基数目 | Polymerase read total bases(bp) | 11 049 130 528 |
| 下机的reads数目 | Number of polymerase reads | 531 175 |
| 下机的read平均长度 | Post-filter mean read length | 20 801 |
| 下机的read的N50长度 | Polymerase read N50 | 35 250 |
| 去掉接头后的碱基数目 | Subreads total bases(bp) | 11 029 483 296 |
| 去掉接头后的 Subread数目 | Number of subreads | 970 426 |
| 去掉接头后的 Subread 平均长度 | Mean subread length(bp) | 11 365.61 |
| 去掉接头后的 Subread N50长度 | Subreads N50 length | 15 985 |
| 过滤无效数据后Subreads总长度 | Subreads total bases(bp) | 10 997 790 898 |
| 过滤无效数据后Subreads数目 | Number of subreads | 908 820 |
| 过滤无效数据后Subreads平均长度* | Subreads mean length(bp)* | 12 101 |
| 过滤无效数据后Subreads N50长度 | Subreads N50 length(bp) | 16 018* |
Table 2 Statistics of third generation sequencing data and filtering results
| 项目 Item | 数据 Data | |
|---|---|---|
| 下机的碱基数目 | Polymerase read total bases(bp) | 11 049 130 528 |
| 下机的reads数目 | Number of polymerase reads | 531 175 |
| 下机的read平均长度 | Post-filter mean read length | 20 801 |
| 下机的read的N50长度 | Polymerase read N50 | 35 250 |
| 去掉接头后的碱基数目 | Subreads total bases(bp) | 11 029 483 296 |
| 去掉接头后的 Subread数目 | Number of subreads | 970 426 |
| 去掉接头后的 Subread 平均长度 | Mean subread length(bp) | 11 365.61 |
| 去掉接头后的 Subread N50长度 | Subreads N50 length | 15 985 |
| 过滤无效数据后Subreads总长度 | Subreads total bases(bp) | 10 997 790 898 |
| 过滤无效数据后Subreads数目 | Number of subreads | 908 820 |
| 过滤无效数据后Subreads平均长度* | Subreads mean length(bp)* | 12 101 |
| 过滤无效数据后Subreads N50长度 | Subreads N50 length(bp) | 16 018* |
| Property | Min | Max | Min | Max | Min | Max |
|---|---|---|---|---|---|---|
| Heterozygosity/% | 1.965 62 | 2.008 05 | 1.955 13 | 1.973 2 | 1.958 25 | 1.984 25 |
| Genome haploid length/ bp | 46 347 904 | 46 440 807 | 50 222 379 | 50 301 292 | 48 878 189 | 48 958 199 |
| Genome repeat length/ bp | 4 637 105 | 4 646 400 | 8 024 318 | 8 036 926 | 6 841 040 | 6 852 239 |
| Sequences repeat rate/% | 10.005 | 10.005 | 15.977 | 15.977 | 13.996 | 13.996 |
| Genome unique length/ bp | 41 710 799 | 41 794 407 | 42 198 061 | 42 264 366 | 42 037 149 | 42 105 960 |
| Model fit/% | 95.5952 | NA | 96.9219 | 98.2063 | 96.2289 | 96.9645 |
| Read error rate/% | 0.253032 | 0.253032 | 0.229964 | 0.229964 | 0.238092 | 0.238092 |
Table 3 Assessment of genome size and heterozygosity(k = 17)
| Property | Min | Max | Min | Max | Min | Max |
|---|---|---|---|---|---|---|
| Heterozygosity/% | 1.965 62 | 2.008 05 | 1.955 13 | 1.973 2 | 1.958 25 | 1.984 25 |
| Genome haploid length/ bp | 46 347 904 | 46 440 807 | 50 222 379 | 50 301 292 | 48 878 189 | 48 958 199 |
| Genome repeat length/ bp | 4 637 105 | 4 646 400 | 8 024 318 | 8 036 926 | 6 841 040 | 6 852 239 |
| Sequences repeat rate/% | 10.005 | 10.005 | 15.977 | 15.977 | 13.996 | 13.996 |
| Genome unique length/ bp | 41 710 799 | 41 794 407 | 42 198 061 | 42 264 366 | 42 037 149 | 42 105 960 |
| Model fit/% | 95.5952 | NA | 96.9219 | 98.2063 | 96.2289 | 96.9645 |
| Read error rate/% | 0.253032 | 0.253032 | 0.229964 | 0.229964 | 0.238092 | 0.238092 |
| 数据库 Database | *版本或公开时间 Version or publication time |
|---|---|
| Gene Ontology(GO) | Releases_2017-09-08 |
| Kyoto Encyclopedia of Genes and Genomes,KEGG | v_81 |
| Cluster of Orthologous Groups of proteins,COG | Releases_2014-11-10 |
| Swiss-Pro | Release_2017-07 |
| Trembl | Release_2017-09 |
| NR | Release_2017-10-10 |
| EggNOG | v_4.5 |
| Antibiotic Resistance Genes Database,ARDB | v_1.1 |
| Pathogen Host Interactions,PHI | v_4.3 |
| Fungal Cytochrome P450 Database | v_1.1 |
| Carbohydrate-Active enzymes Database,CAZy | Release_2017-09 |
| Virulence Factor Database,VFDB | Release_2017-09 |
| Type III Secretion System Effector Proteins,T3SS | v_1.0 |
| TransportDB | v_2.0 |
Table 4 Protein database for genome annotation
| 数据库 Database | *版本或公开时间 Version or publication time |
|---|---|
| Gene Ontology(GO) | Releases_2017-09-08 |
| Kyoto Encyclopedia of Genes and Genomes,KEGG | v_81 |
| Cluster of Orthologous Groups of proteins,COG | Releases_2014-11-10 |
| Swiss-Pro | Release_2017-07 |
| Trembl | Release_2017-09 |
| NR | Release_2017-10-10 |
| EggNOG | v_4.5 |
| Antibiotic Resistance Genes Database,ARDB | v_1.1 |
| Pathogen Host Interactions,PHI | v_4.3 |
| Fungal Cytochrome P450 Database | v_1.1 |
| Carbohydrate-Active enzymes Database,CAZy | Release_2017-09 |
| Virulence Factor Database,VFDB | Release_2017-09 |
| Type III Secretion System Effector Proteins,T3SS | v_1.0 |
| TransportDB | v_2.0 |
| 基因组Genome | 序列类型 Seq type | 数目 Total number | 组装总长度 Total length/bp | 序列N50长度 N50 length/bp | 间隙数 Gap number | 序列GC含量 GC content /% |
|---|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Scaffold | 71 | 56 763 274 | 1 200 238 | 0 | 56.14 |
| Contig | 71 | 56 763 274 | 1 200 238 | - | 56.14 | |
| 线粒体基因组Mitochondrion genome | Scaffold | 1 | 85 353 | 85 353 | 0 | 26.91 |
| Contig | 1 | 85 353 | 85 353 | - | 26.91 |
Table 5 Whole genome of Ganoderma sp. strain Zizhi S2* assembled by De novo sequencing
| 基因组Genome | 序列类型 Seq type | 数目 Total number | 组装总长度 Total length/bp | 序列N50长度 N50 length/bp | 间隙数 Gap number | 序列GC含量 GC content /% |
|---|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Scaffold | 71 | 56 763 274 | 1 200 238 | 0 | 56.14 |
| Contig | 71 | 56 763 274 | 1 200 238 | - | 56.14 | |
| 线粒体基因组Mitochondrion genome | Scaffold | 1 | 85 353 | 85 353 | 0 | 26.91 |
| Contig | 1 | 85 353 | 85 353 | - | 26.91 |
| Assembly length/bp | Coverage(#) | Coverage rate/% | Depth(#) | Reads usage percent/% |
|---|---|---|---|---|
| 56 848 627 | 56 499 028 | 99.39 | 177 | 83.45 |
Table 6 Genome sequencing depth and coverage
| Assembly length/bp | Coverage(#) | Coverage rate/% | Depth(#) | Reads usage percent/% |
|---|---|---|---|---|
| 56 848 627 | 56 499 028 | 99.39 | 177 | 83.45 |
| 基因组 Genome | 类型 Type | 数目 Total number | 序列总长度 Total length/bp | 平均长度 Average length/bp | 占基因组总长的比例 Percentage in total length /% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Gene | 16 681 | 32 438 896 | 1 944.66 | 57.15 |
| Exons | 95 839 | 23 930 217 | 249.69 | 42.16 | |
| CDS | 16 681 | 23 930 217 | 1 434.58 | 42.16 | |
| Introne | 79 158 | 8 508 679 | 107.49 | 14.99 | |
| 线粒体基因组 Mitochondrion genome | Gene | 56 | 77 918 | 1 391.39 | 91.29 |
| Exons | 78 | 46 571 | 597.06 | 54.56 | |
| CDS | 56 | 46 571 | 831.62 | 54.56 | |
| Introne | 22 | 29 854 | 1 357.00 | 34.98 |
Table 7 Statistics of gene composition of the genome
| 基因组 Genome | 类型 Type | 数目 Total number | 序列总长度 Total length/bp | 平均长度 Average length/bp | 占基因组总长的比例 Percentage in total length /% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | Gene | 16 681 | 32 438 896 | 1 944.66 | 57.15 |
| Exons | 95 839 | 23 930 217 | 249.69 | 42.16 | |
| CDS | 16 681 | 23 930 217 | 1 434.58 | 42.16 | |
| Introne | 79 158 | 8 508 679 | 107.49 | 14.99 | |
| 线粒体基因组 Mitochondrion genome | Gene | 56 | 77 918 | 1 391.39 | 91.29 |
| Exons | 78 | 46 571 | 597.06 | 54.56 | |
| CDS | 56 | 46 571 | 831.62 | 54.56 | |
| Introne | 22 | 29 854 | 1 357.00 | 34.98 |
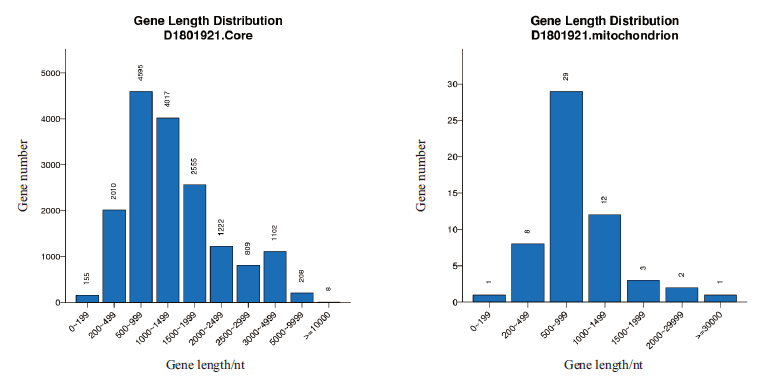
Fig. 2 Gene length distribution in nuclear genome and mitochondrial genome The abscissa is gene length,and the ordinate is the number of genes corresponding to the gene length
| Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number |
|---|---|---|---|---|---|---|---|---|
| Scaffold_1 | 4469875 | 1393 | Scaffold_31 | 666660 | 203 | Scaffold_53 | 207498 | 52 |
| Scaffold_10 | 1494552 | 424 | Scaffold_32 | 647059 | 167 | Scaffold_54 | 183478 | 51 |
| Scaffold_11 | 1421539 | 426 | Scaffold_33 | 627493 | 161 | Scaffold_55 | 176875 | 41 |
| Scaffold_12 | 1200238 | 334 | Scaffold_34 | 621976 | 186 | Scaffold_56 | 174271 | 58 |
| Scaffold_13 | 1081736 | 337 | Scaffold_35 | 614529 | 155 | Scaffold_57 | 167193 | 47 |
| Scaffold_14 | 1072049 | 347 | Scaffold_36 | 532656 | 153 | Scaffold_58 | 161880 | 47 |
| Scaffold_15 | 1050642 | 314 | Scaffold_37 | 511480 | 126 | Scaffold_59 | 161526 | 43 |
| Scaffold_16 | 1046259 | 337 | Scaffold_38 | 506477 | 122 | Scaffold_6 | 2277588 | 668 |
| Scaffold_17 | 1019206 | 303 | Scaffold_39 | 493120 | 139 | Scaffold_60 | 146901 | 39 |
| Scaffold_18 | 950046 | 247 | Scaffold_4 | 3153885 | 1014 | Scaffold_61 | 115424 | 31 |
| Scaffold_19 | 926904 | 298 | Scaffold_40 | 428051 | 134 | Scaffold_62 | 114060 | 27 |
| Scaffold_2 | 3484778 | 1149 | Scaffold_41 | 415410 | 111 | Scaffold_63 | 103079 | 35 |
| Scaffold_20 | 895893 | 264 | Scaffold_42 | 375402 | 111 | Scaffold_64 | 95381 | 24 |
| Scaffold_21 | 879786 | 257 | Scaffold_43 | 333682 | 104 | Scaffold_65 | 86665 | 14 |
| Scaffold_22 | 879764 | 234 | Scaffold_44 | 331534 | 85 | Scaffold_66 | 83587 | 23 |
| Scaffold_23 | 848793 | 236 | Scaffold_45 | 319851 | 90 | Scaffold_67 | 78814 | 27 |
| Scaffold_24 | 842024 | 258 | Scaffold_46 | 311630 | 87 | Scaffold_68 | 66934 | 13 |
| Scaffold_25 | 790967 | 201 | Scaffold_47 | 282820 | 90 | Scaffold_69 | 59421 | 15 |
| Scaffold_26 | 790090 | 218 | Scaffold_48 | 257692 | 74 | Scaffold_7 | 2147372 | 600 |
| Scaffold_27 | 747333 | 216 | Scaffold_49 | 248,648 | 51 | Scaffold_70 | 58166 | 16 |
| Scaffold_28 | 714016 | 198 | Scaffold_5 | 2522070 | 731 | Scaffold_71 | 33177 | 15 |
| Scaffold_29 | 692498 | 211 | Scaffold_50 | 232229 | 58 | Scaffold_72 | 85353 | 56 |
| Scaffold_3 | 3315994 | 946 | Scaffold_51 | 226670 | 86 | Scaffold_8 | 2100477 | 654 |
| Scaffold_30 | 691292 | 185 | Scaffold_52 | 212908 | 70 | Scaffold_9 | 1783301 | 500 |
Table 8 Statistics on the total sequence length and genes’ number on each scaffold
| Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number | Seq_name | Seq_length | Gene number |
|---|---|---|---|---|---|---|---|---|
| Scaffold_1 | 4469875 | 1393 | Scaffold_31 | 666660 | 203 | Scaffold_53 | 207498 | 52 |
| Scaffold_10 | 1494552 | 424 | Scaffold_32 | 647059 | 167 | Scaffold_54 | 183478 | 51 |
| Scaffold_11 | 1421539 | 426 | Scaffold_33 | 627493 | 161 | Scaffold_55 | 176875 | 41 |
| Scaffold_12 | 1200238 | 334 | Scaffold_34 | 621976 | 186 | Scaffold_56 | 174271 | 58 |
| Scaffold_13 | 1081736 | 337 | Scaffold_35 | 614529 | 155 | Scaffold_57 | 167193 | 47 |
| Scaffold_14 | 1072049 | 347 | Scaffold_36 | 532656 | 153 | Scaffold_58 | 161880 | 47 |
| Scaffold_15 | 1050642 | 314 | Scaffold_37 | 511480 | 126 | Scaffold_59 | 161526 | 43 |
| Scaffold_16 | 1046259 | 337 | Scaffold_38 | 506477 | 122 | Scaffold_6 | 2277588 | 668 |
| Scaffold_17 | 1019206 | 303 | Scaffold_39 | 493120 | 139 | Scaffold_60 | 146901 | 39 |
| Scaffold_18 | 950046 | 247 | Scaffold_4 | 3153885 | 1014 | Scaffold_61 | 115424 | 31 |
| Scaffold_19 | 926904 | 298 | Scaffold_40 | 428051 | 134 | Scaffold_62 | 114060 | 27 |
| Scaffold_2 | 3484778 | 1149 | Scaffold_41 | 415410 | 111 | Scaffold_63 | 103079 | 35 |
| Scaffold_20 | 895893 | 264 | Scaffold_42 | 375402 | 111 | Scaffold_64 | 95381 | 24 |
| Scaffold_21 | 879786 | 257 | Scaffold_43 | 333682 | 104 | Scaffold_65 | 86665 | 14 |
| Scaffold_22 | 879764 | 234 | Scaffold_44 | 331534 | 85 | Scaffold_66 | 83587 | 23 |
| Scaffold_23 | 848793 | 236 | Scaffold_45 | 319851 | 90 | Scaffold_67 | 78814 | 27 |
| Scaffold_24 | 842024 | 258 | Scaffold_46 | 311630 | 87 | Scaffold_68 | 66934 | 13 |
| Scaffold_25 | 790967 | 201 | Scaffold_47 | 282820 | 90 | Scaffold_69 | 59421 | 15 |
| Scaffold_26 | 790090 | 218 | Scaffold_48 | 257692 | 74 | Scaffold_7 | 2147372 | 600 |
| Scaffold_27 | 747333 | 216 | Scaffold_49 | 248,648 | 51 | Scaffold_70 | 58166 | 16 |
| Scaffold_28 | 714016 | 198 | Scaffold_5 | 2522070 | 731 | Scaffold_71 | 33177 | 15 |
| Scaffold_29 | 692498 | 211 | Scaffold_50 | 232229 | 58 | Scaffold_72 | 85353 | 56 |
| Scaffold_3 | 3315994 | 946 | Scaffold_51 | 226670 | 86 | Scaffold_8 | 2100477 | 654 |
| Scaffold_30 | 691292 | 185 | Scaffold_52 | 212908 | 70 | Scaffold_9 | 1783301 | 500 |
| 基因组 Genome | 类型 Type | 拷贝数目 Number of copies | 平均长度 Average length/bp | 总长度 Total length/bp | 占基因组百分比 Percentage in genome/% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | tRNA | 226 | 81.04 | 18 316 | 0.0323 |
| rRNA(by De novo prediction)* | 12 | 2 883.08 | 34 597 | 0.0609 | |
| sRNA | 64 | 62.23 | 3 983 | 0.007 | |
| snRNA | 29 | 102.79 | 2 981 | 0.0053 | |
| miRNA | 74 | 59.91 | 4 434 | 0.0078 | |
| 线粒体基因组 Mitochondrion genome | tRNA | 20 | 72.9 | 1 458 | 1.7082 |
| rRNA(by De novo prediction)** | 0 | 0 | 0 | 0 | |
| sRNA | 1 | 90 | 90 | 0.1054 | |
| snRNA | 1 | 38 | 38 | 0.0445 | |
| miRNA | 1 | 88 | 88 | 0.1031 |
Table 9 Statistics of non-coding RNA
| 基因组 Genome | 类型 Type | 拷贝数目 Number of copies | 平均长度 Average length/bp | 总长度 Total length/bp | 占基因组百分比 Percentage in genome/% |
|---|---|---|---|---|---|
| 核基因组 Nuclear genome | tRNA | 226 | 81.04 | 18 316 | 0.0323 |
| rRNA(by De novo prediction)* | 12 | 2 883.08 | 34 597 | 0.0609 | |
| sRNA | 64 | 62.23 | 3 983 | 0.007 | |
| snRNA | 29 | 102.79 | 2 981 | 0.0053 | |
| miRNA | 74 | 59.91 | 4 434 | 0.0078 | |
| 线粒体基因组 Mitochondrion genome | tRNA | 20 | 72.9 | 1 458 | 1.7082 |
| rRNA(by De novo prediction)** | 0 | 0 | 0 | 0 | |
| sRNA | 1 | 90 | 90 | 0.1054 | |
| snRNA | 1 | 38 | 38 | 0.0445 | |
| miRNA | 1 | 88 | 88 | 0.1031 |
| Genome | Method | Repeat size/bp | Percentage in genome/% |
|---|---|---|---|
| 核基因组 Nuclear genome | Repbase | 1 307 464 | 2.3034 |
| ProMask | 1 973 669 | 3.477 | |
| De novo | 6 991 719 | 12.3173 | |
| TRF | 326 000 | 0.5743 | |
| Total | 7 954 327 | 14.0132 | |
| 线粒体基因组 Mitochondrion genome | Repbase | 128 | 0.15 |
| ProMask | 1 005 | 1.1775 | |
| De novo | 0 | ||
| TRF | 320 | 0.3749 | |
| Total | 1 453 | 1.7023 |
Table 10 Statistics of repeat sequences in the genome
| Genome | Method | Repeat size/bp | Percentage in genome/% |
|---|---|---|---|
| 核基因组 Nuclear genome | Repbase | 1 307 464 | 2.3034 |
| ProMask | 1 973 669 | 3.477 | |
| De novo | 6 991 719 | 12.3173 | |
| TRF | 326 000 | 0.5743 | |
| Total | 7 954 327 | 14.0132 | |
| 线粒体基因组 Mitochondrion genome | Repbase | 128 | 0.15 |
| ProMask | 1 005 | 1.1775 | |
| De novo | 0 | ||
| TRF | 320 | 0.3749 | |
| Total | 1 453 | 1.7023 |
| Genome | Method | Type | DNA | LINE | LTR | SINE | Other | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| 核基因组Nuclear genome | Repbase TEs | Length(bp) | 95 474 | 66 559 | 1 146 867 | 2 711 | 165 | 1 360 | 1 307 464 |
| % in Genome | 0.1682 | 0.1173 | 2.0204 | 0.0048 | 0.0003 | 0.0024 | 2.3034 | ||
| ProteinMask TEs | Length(bp) | 316 367 | 100 469 | 1 558 079 | 0 | 0 | 0 | 1 973 669 | |
| % in Genome | 0.5573 | 0.177 | 2.7449 | 0 | 0 | 0 | 3.477 | ||
| De novo TEs | Length(bp) | 594 332 | 431 488 | 2 449 588 | 4 081 | 0 | 3 592 915 | 6 991 719 | |
| % in Genome | 1.047 | 0.7602 | 4.3154 | 0.0072 | 0 | 6.3296 | 12.3173 | ||
| Combined TEs | Length(bp) | 924 792 | 515 460 | 2 849 776 | 5 828 | 165 | 3 594 275 | 7 718 181 | |
| % in Genome | 1.6292 | 0.9081 | 5.0205 | 0.0103 | 0.0003 | 6.332 | 13.5971 | ||
| 线粒体基因组 Mitochondrion genome | Repbase TEs | Length(bp) | 0 | 0 | 128 | 0 | 0 | 0 | 128 |
| % in Genome | 0 | 0 | 0.15 | 0 | 0 | 0 | 0.15 | ||
| ProteinMask TEs | Length(bp) | 0 | 1 005 | 0 | 0 | 0 | 0 | 1 005 | |
| % in Genome | 0 | 1.1775 | 0 | 0 | 0 | 0 | 1.1775 | ||
| De novo TEs | Length(bp) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| % in Genome | - | - | - | - | - | - | - | ||
| Combined TEs | Length(bp) | 0 | 1 005 | 128 | 0 | 0 | 0 | 1 133 | |
| % in Genome | 0 | 1.1775 | 0.15 | 0 | 0 | 0 | 1.3274 |
Table 11 Statistics of transposable factors
| Genome | Method | Type | DNA | LINE | LTR | SINE | Other | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| 核基因组Nuclear genome | Repbase TEs | Length(bp) | 95 474 | 66 559 | 1 146 867 | 2 711 | 165 | 1 360 | 1 307 464 |
| % in Genome | 0.1682 | 0.1173 | 2.0204 | 0.0048 | 0.0003 | 0.0024 | 2.3034 | ||
| ProteinMask TEs | Length(bp) | 316 367 | 100 469 | 1 558 079 | 0 | 0 | 0 | 1 973 669 | |
| % in Genome | 0.5573 | 0.177 | 2.7449 | 0 | 0 | 0 | 3.477 | ||
| De novo TEs | Length(bp) | 594 332 | 431 488 | 2 449 588 | 4 081 | 0 | 3 592 915 | 6 991 719 | |
| % in Genome | 1.047 | 0.7602 | 4.3154 | 0.0072 | 0 | 6.3296 | 12.3173 | ||
| Combined TEs | Length(bp) | 924 792 | 515 460 | 2 849 776 | 5 828 | 165 | 3 594 275 | 7 718 181 | |
| % in Genome | 1.6292 | 0.9081 | 5.0205 | 0.0103 | 0.0003 | 6.332 | 13.5971 | ||
| 线粒体基因组 Mitochondrion genome | Repbase TEs | Length(bp) | 0 | 0 | 128 | 0 | 0 | 0 | 128 |
| % in Genome | 0 | 0 | 0.15 | 0 | 0 | 0 | 0.15 | ||
| ProteinMask TEs | Length(bp) | 0 | 1 005 | 0 | 0 | 0 | 0 | 1 005 | |
| % in Genome | 0 | 1.1775 | 0 | 0 | 0 | 0 | 1.1775 | ||
| De novo TEs | Length(bp) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| % in Genome | - | - | - | - | - | - | - | ||
| Combined TEs | Length(bp) | 0 | 1 005 | 128 | 0 | 0 | 0 | 1 133 | |
| % in Genome | 0 | 1.1775 | 0.15 | 0 | 0 | 0 | 1.3274 |
| 数据库 | Total | P450 | TF | CAZY | PHI | IPR |
|---|---|---|---|---|---|---|
| Nuclear genome | 16 681 | 1 261(7.55%) | 588(3.52%) | 278(1.66%) | 570(3.41%) | 10 296(61.72%) |
| Mitochondrial genome | 56 | 0(0%) | 0(0%) | 0(0%) | 1(1.78%) | 47(83.92%) |
| 数据库 | SWISSPROT | COG | GO | KEGG | NR | Over All |
| Nuclear genome | 2 708(16.23%) | 1 244(7.45%) | 7 373(44.19%) | 4 346(26.05%) | 12 161(72.9%) | 13 161(78.89%) |
| Mitochondrial genome | 24(42.85%) | 10(17.85%) | 44(78.57%) | 16(28.57%) | 46(82.14%) | 51(91.07%) |
Table 12 Statistics of gene set annotation results
| 数据库 | Total | P450 | TF | CAZY | PHI | IPR |
|---|---|---|---|---|---|---|
| Nuclear genome | 16 681 | 1 261(7.55%) | 588(3.52%) | 278(1.66%) | 570(3.41%) | 10 296(61.72%) |
| Mitochondrial genome | 56 | 0(0%) | 0(0%) | 0(0%) | 1(1.78%) | 47(83.92%) |
| 数据库 | SWISSPROT | COG | GO | KEGG | NR | Over All |
| Nuclear genome | 2 708(16.23%) | 1 244(7.45%) | 7 373(44.19%) | 4 346(26.05%) | 12 161(72.9%) | 13 161(78.89%) |
| Mitochondrial genome | 24(42.85%) | 10(17.85%) | 44(78.57%) | 16(28.57%) | 46(82.14%) | 51(91.07%) |
| Species and strain name | G. sp. Zizhi S2 | G. lucidum G.260125-1 | G. sinense ZZ0214-1 |
|---|---|---|---|
| Chromosomes | NA* | 13 | 12 |
| Number of scaffolds | 71 | 82 | 69 |
| Length of genome assembly(bp) | 56 763 274 | 43 292 570 | 48 955 697 |
| GC content(%) | 56.14 | 56.16 | 56.17 |
| Number of protein-coding genes | 16 681 | 16 495 | 15 688 |
| Average gene length(bp) | 1 944.66 | 1 569.02 | 1 668.82 |
| GC content of protein-coding genes(%) | 59.62 | 59.33 | 59.53 |
| Average number of exons per gene | 5.74 | 4.58 | 5.34 |
| Average exon size(bp) | 249.69 | 259 | 245.67 |
| Average coding sequence size(bp) | 1 434.57 | 1 188.79 | 1 313.03 |
| Average intron size(bp) | 107.24 | 94.12 | 81.71 |
| Average size of intergenic regions(bp) | 1 452.02 | 1 050.52 | 1 487.12 |
| Repeat sequences(%) | 14.01 | 8.15 | 8.53 |
| Sequencing platform | PacBio Sequel & Illumina HiSeq | Roche 450 & Illumina | Roche 454 & Illumina HiSeq |
Table 13 Information comparison of 3 nuclear genomes of Ganoderma spp.
| Species and strain name | G. sp. Zizhi S2 | G. lucidum G.260125-1 | G. sinense ZZ0214-1 |
|---|---|---|---|
| Chromosomes | NA* | 13 | 12 |
| Number of scaffolds | 71 | 82 | 69 |
| Length of genome assembly(bp) | 56 763 274 | 43 292 570 | 48 955 697 |
| GC content(%) | 56.14 | 56.16 | 56.17 |
| Number of protein-coding genes | 16 681 | 16 495 | 15 688 |
| Average gene length(bp) | 1 944.66 | 1 569.02 | 1 668.82 |
| GC content of protein-coding genes(%) | 59.62 | 59.33 | 59.53 |
| Average number of exons per gene | 5.74 | 4.58 | 5.34 |
| Average exon size(bp) | 249.69 | 259 | 245.67 |
| Average coding sequence size(bp) | 1 434.57 | 1 188.79 | 1 313.03 |
| Average intron size(bp) | 107.24 | 94.12 | 81.71 |
| Average size of intergenic regions(bp) | 1 452.02 | 1 050.52 | 1 487.12 |
| Repeat sequences(%) | 14.01 | 8.15 | 8.53 |
| Sequencing platform | PacBio Sequel & Illumina HiSeq | Roche 450 & Illumina | Roche 454 & Illumina HiSeq |
| [1] | 钟礼义. 紫芝新品种武芝2号区域试验[J]. 中国食用菌, 2013, 32(6): 25-27. |
| Zhong LY. The regional test on new varieties of Ganoderma[J]. Edible Fungi China, 2013, 32(6): 25-27. | |
| [2] | 钟礼义, 陈体强, 刘新锐, 等. 鉴别紫芝菌株的PCR引物筛选及其序列比对验证[J]. 福建农业学报, 2020, 35(7): 725-730. |
| Zhong LY, Chen TQ, Liu XR, et al. Selection and sequence alignment of PCR primers for identifying Zizhi strain[J]. Fujian J Agric Sci, 2020, 35(7): 725-730. | |
| [3] | 陈体强, 吴锦忠, 钟礼义, 等. 福建野生紫芝资源的开发利用Ⅰ. 硬孔灵芝[J]. 福建农业大学学报:自然科学版, 2006, 35(3): 324-328. |
| Chen TQ, Wu JZ, Zhong LY, et al. Exploitation and utilization of wild Zhizi resource in Fujian Ⅰ. Ganoderma duropora(Sect. Phaeonema)[J]. J Fujian Agric For Univ:Nat Sci Edn, 2006, 35(3): 324-328. | |
| [4] | Hapuarachchi KK, Karunarathna SC, McKenzie EHC, et al. High phenotypic plasticity of Ganoderma sinense(Ganodermataceae, Polyporales)in China[J]. Asian J Mycol, 2019, 2(1): 1-47. |
| [5] | Wang DM, Zhang XQ, Yao YJ. Type studies of some Ganoderma species from China[J]. Mycotaxon, 2005, 93: 61-70. |
| [6] | 王冬梅. 中国灵芝属系统发育研究[D]. 北京:中国科学院微生物研究所, 2005. |
| Wang DM. Phylogeny of Ganoderma in China[D]. Beijing:Institute of Microbiology, Chinese Academy of Sciences, 2005. | |
| [7] | 曹云. 中国灵芝属的系统学研究[D]. 北京:中国科学院大学, 2013. |
| Cao Y. Systematic study of Ganoderma in China[D]. Beijing:University of Chinese Academy of Sciences, 2013. | |
| [8] | 王新存. 灵芝科系统发育研究[D]. 北京:中国科学院大学, 2012. |
| Wang XC. Phylogenetic studies of Ganodermataceae. Beijing:University of Chinese Academy of Sciences, 2012. | |
| [9] | 邢佳慧. 灵芝属的物种多样性、分类与系统发育研究[D]. 北京:北京林业大学, 2019. |
| Xing JH. Species diversity, taxonomy and phylogeny of Ganoderma[D]. Beijing:Beijing Forestry University, 2019. | |
| [10] | 陈体强, 吴锦忠, 李晔, 等. 福建野生紫芝资源开发利用Ⅱ. ‘闽紫96’(中国灵芝)[J]. 菌物研究, 2006, 4(4): 27-32. |
| Chen TQ, Wu JZ, Li Y, et al. Exploitation and utilization of wild Zhizi resources(Sect. Phaeonema of Ganoderma)in Fujian(Ⅱ). G. sinense ‘minzi 96’[J]. J Fungal Res, 2006, 4(4): 27-32. | |
| [11] |
Kim KE, Peluso P, Babayan P, et al. Long-read, whole-genome shotgun sequence data for five model organisms[J]. Sci Data, 2014, 1: 140045.
doi: 10.1038/sdata.2014.45 URL |
| [12] | Faino L, Seidl MF, Datema E, et al. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome[J]. mBio, 2015, 6(4): e00936-15. |
| [13] |
Sit CS, Ruzzini AC, Van Arnam EB, et al. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants[J]. PNAS, 2015, 112(43): 13150-13154.
doi: 10.1073/pnas.1515348112 URL |
| [14] | Tsuji M, Kudoh S, Hoshino T. Draft genome sequence of cryophilic basidiomycetous yeast Mrakia blollopis SK-4, isolated from an algal mat of Naga-ike Lake in the Skarvsnes ice-free area, East Antarctica[J]. Genome Announc, 2015, 3(1): e01454-14. |
| [15] |
Badouin H, Hood ME, Gouzy J, et al. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae[J]. Genetics, 2015, 200(4): 1275-1284.
doi: 10.1534/genetics.115.177709 pmid: 26044594 |
| [16] |
Fei X, Zhao MW, Li YX. Cloning and sequence analysis of a glyceraldehyde-3-phosphate dehydrogenase gene from Ganoderma lucidum[J]. J Microbiol, 2006, 44(5): 515-522.
pmid: 17082745 |
| [17] | 张妍, 黄晨阳, 高巍. 食用菌分子育种研究进展[J]. 菌物研究, 2019, 17(4): 229-239. |
| Zhang Y, Huang CY, Gao W. Research advances on molecular mushroom breeding[J]. J Fungal Res, 2019, 17(4): 229-239. | |
| [18] | 左斌, 王海斌, 葛俊, 等. 真菌漆酶基因研究进展[J]. 微生物学免疫学进展, 2009, 37(1): 72-76. |
| Zuo B, Wang HB, Ge J, et al. Research progress in the gene of fungal laccase[J]. Prog Microbiol Immunol, 2009, 37(1): 72-76. | |
| [19] |
Sun SJ, Liu JZ, Hu KH, et al. The level of secreted laccase activity in the edible fungi and their growing cycles are closely related[J]. Curr Microbiol, 2011, 62(3): 871-875.
doi: 10.1007/s00284-010-9794-z pmid: 21046396 |
| [20] | 陈体强, 吴建国, 毛方华, 等. 硬孔灵芝和赤芝孢子油的脂肪酸、角鲨烯及麦角甾醇类的比较[J]. 食用菌学报, 2018, 25(1): 47-52. |
| Chen TQ, Wu JG, Mao FH, et al. Comparison on fatty acid profile, squalene and ergosterols between spore oils extracted from Ganoderma duropora and G. lucidum[J]. Acta Edulis Fungi, 2018, 25(1): 47-52. | |
| [21] |
Chen TQ, Wu YB, Wu JG, et al. Fatty acids, essential oils, and squalene in the spore lipids of Ganoderma lucidum by GC-MS and GC-FID[J]. Chem Nat Compd, 2013, 49(1): 143-144.
doi: 10.1007/s10600-013-0536-x URL |
| [22] |
Lian CL, Wu YQ, Chen TQ, et al. Identification of new trace triterpenoids from the fungus Ganoderma duripora[J]. Phytochem Lett, 2017, 21: 237-239.
doi: 10.1016/j.phytol.2017.07.005 URL |
| [23] |
Chen TQ, Wu JG, Wu YB, et al. Supercritical fluid CO2 extraction, simultaneous determination of total sterols in the spore lipids of Ganoderma lucidum by GC-MS/SIM methods[J]. Chem Nat Compd, 2012, 48(4): 657-658.
doi: 10.1007/s10600-012-0338-6 URL |
| [24] | 赵芬, 李晔, 刘超, 等. 硬孔灵芝的化学成分研究[J]. 菌物学报, 2009, 28(3): 407-409. |
| Zhao F, Li Y, Liu C, et al. Chemical components from the fruiting bodies of Ganoderma duropora[J]. Mycosystema, 2009, 28(3): 407-409. | |
| [25] |
Lian CL, Wang CF, Xiao Q, et al. The triterpenes and steroids from the fruiting body Ganoderma duripora[J]. Biochem Syst Ecol, 2017, 73: 50-53.
doi: 10.1016/j.bse.2017.06.005 URL |
| [26] | 方星, 师亮, 徐颖洁, 等. 灵芝甾醇14α-脱甲基酶基因的克隆及超量表达对三萜合成的影响[J]. 菌物学报, 2011, 30(2): 242-248. |
| Fang X, Shi L, Xu YJ, et al. Cloning of a sterol 14α-demethylase gene and the effects of over-expression of the gene on biological synjournal of triterpenes in Ganoderma lucidum[J]. Mycosystema, 2011, 30(2): 242-248. | |
| [27] | Grigoriev IV, Dullen D, Goodwin SB, et al. Fueling the future with fungal genomics[J]. Mycology, 2011, 2(3): 192-209. |
| [28] |
Grigoriev IV, Nikitin R, Haridas S, et al. MycoCosm portal:gearing up for 1000 fungal genomes[J]. Nucleic Acids Res, 2014, 42(database issue): D699-D704.
doi: 10.1093/nar/gkt1183 URL |
| [29] | 刘淑艳, 李广, 李玉. 菌物基因组测定研究进展[J]. 吉林农业大学学报, 2014, 36(1): 1-9, 16. |
| Liu SY, Li G, Li Y. Advance of fungal genomes sequencing researches[J]. J Jilin Agric Univ, 2014, 36(1): 1-9, 16. | |
| [30] | 凌志琳, 赵瑞琳. 基因组学在食药用菌栽培育种中的研究进展[J]. 食用菌学报, 2018, 25(1): 93-106. |
| Ling ZL, Zhao RL. Advances of genomics-assisted cultivation and breeding of edible and medicinal mushrooms[J]. Acta Edulis Fungi, 2018, 25(1): 93-106. | |
| [31] |
Chen SL, Xu J, Liu C, et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum[J]. Nat Commun, 2012, 3: 913.
doi: 10.1038/ncomms1923 URL |
| [32] |
Zhu YJ, Xu J, Sun C, et al. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense[J]. Sci Rep, 2015, 5: 11087.
doi: 10.1038/srep11087 URL |
| [33] |
Benveniste P. Biosynjournal and accumulation of sterols[J]. Annu Rev Plant Biol, 2004, 55: 429-457.
pmid: 15377227 |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [3] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [4] | LIU Yue-e, XU Tian-jun, CAI Wan-tao, LYU Tian-fang, ZHANG Yong, XUE Hong-he, WANG Rong-huan, ZHAO Jiu-ran. Current Status and Prospects of Maize Super High Yield Research in China [J]. Biotechnology Bulletin, 2023, 39(8): 52-61. |
| [5] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [6] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [7] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [8] | LIU Chuan-he, HE Han, HE Xiu-gu, CHEN Xin, LIU Kai, SHAO Xue-hua, LAI Duo, QIN Jian, ZHUANG Qing-li, KUANG Shi-zi, XIAO Wei-qiang. Physiological and Metabolitic Mechanisms of Different Pineapple Cultivars Responding to Low Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(10): 219-230. |
| [9] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [10] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [11] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [12] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [13] | HAN Zhi-ling, CHEN Qing, LIANG Xiao, WU Chun-ling, LIU Ying, WU Mu-feng, XU Xue-lian. Influence on Expression of Jasmonic Acid Signaling Pathway Gene in Tetranychus urticae Fed on Mite-resistant and Mite-susceptible Cassava Cultivars [J]. Biotechnology Bulletin, 2022, 38(6): 211-220. |
| [14] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [15] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||