








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 205-217.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0447
Previous Articles Next Articles
QIU Yi-bin1,3( ), MA Yan-qin2, SHA Yuan-yuan2, ZHU Yi-fan2, SU Er-zheng1, LEI Peng2, LI Sha2, XU Hong2
), MA Yan-qin2, SHA Yuan-yuan2, ZHU Yi-fan2, SU Er-zheng1, LEI Peng2, LI Sha2, XU Hong2
Received:2021-04-07
Online:2022-02-26
Published:2022-03-09
QIU Yi-bin, MA Yan-qin, SHA Yuan-yuan, ZHU Yi-fan, SU Er-zheng, LEI Peng, LI Sha, XU Hong. Research Progress in Molecular Genetic Manipulation Technology of Bacillus amyloliquefaciens and Its Application[J]. Biotechnology Bulletin, 2022, 38(2): 205-217.
| Plasmid | Promoter | Terminator | Replicon | Reference |
|---|---|---|---|---|
| pUBXC | PxylA | t0 | repB | [ |
| pDR | PHpaII | Tfd | repF | [ |
| pMA5 | PHpaII | Tfd | repB | |
| pNX01 | P43 | Tamy | Ori-rep(p2Sip) | [ |
| pWH1520 | PxylA | pBC16 ori | [ | |
| pKSV7 | P43 | Temperature-sensitive replication from pE194ts | ||
| pNW33N | ctaB、qcr、qox、resD promoter | repB | [ | |
| pLY-3 | Promoter from amyE of B. subtilis 168 | [ | ||
| pLakr | PQ | [ | ||
| pHT01 | Pgrac | Theta replicon | [ |
Table 1 Synthetic biological genetic elements used in B. amyloliquefaciens
| Plasmid | Promoter | Terminator | Replicon | Reference |
|---|---|---|---|---|
| pUBXC | PxylA | t0 | repB | [ |
| pDR | PHpaII | Tfd | repF | [ |
| pMA5 | PHpaII | Tfd | repB | |
| pNX01 | P43 | Tamy | Ori-rep(p2Sip) | [ |
| pWH1520 | PxylA | pBC16 ori | [ | |
| pKSV7 | P43 | Temperature-sensitive replication from pE194ts | ||
| pNW33N | ctaB、qcr、qox、resD promoter | repB | [ | |
| pLY-3 | Promoter from amyE of B. subtilis 168 | [ | ||
| pLakr | PQ | [ | ||
| pHT01 | Pgrac | Theta replicon | [ |
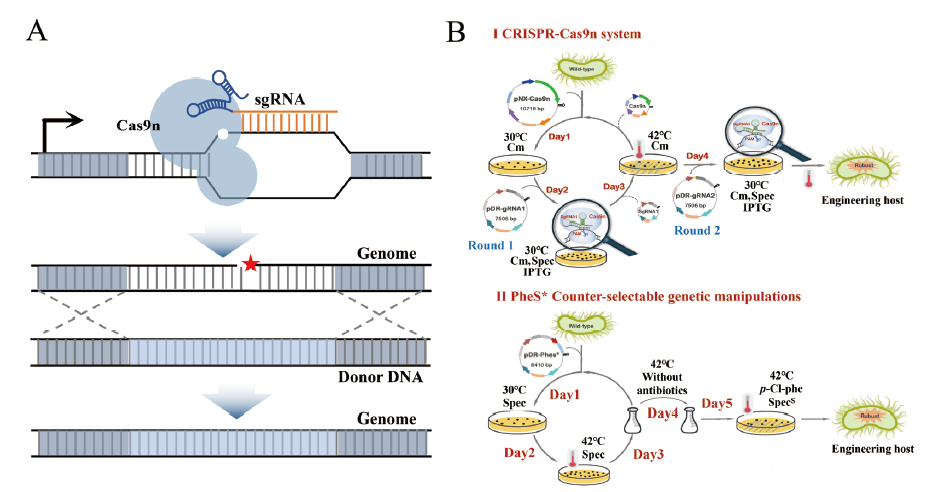
Fig. 2 A schematic diagram of operation while CRISPR-Cas9n gene editing technology in B. amyloliquefaciens A:The gene editing mechanism of CRISPR-Cas9n. B:Comparison of CRISPR-Cas9n and conventional homologous recombination techniques

Fig. 3 Construction of CRISPR-Cas9n gene silencing technology in B. amyloliquefaciens A:The catalytic mechanism of CRISPR-dCas9. B:Schematic representation of the CRISPR-dCas9 double plasmid system. C:Design of sgRNA targeting to egfp expression region. Transcription level of different sgRNA-mediated egfp genes,observed by fluorescence confocal microscopy under different sgRNA regulation
| Strain | Transformation method | Concrete operation | Transformation plasmid | Transformation efficiency /(CFU·μg-1) |
|---|---|---|---|---|
| B. amyloliquefaciens TA208[ | Electroproration | Adding DL-threonine or glycine to weaken the cell wall in hypertonic medium | pUB110 | (1.13 ± 0.34)×107 |
| Adding DL-threonine or glycine to weaken the cell wall in hypertonic medium;heat shock treatment | pHCMC02 | (8.94 ± 0.77)×105 | ||
| B. amyloliquefaciens NB[ | Electroproration | Demethylation | pDR | 4.94 ± 0.42)×104 |
| Demethylation | pMA5 | (6.15 ± 0.19)×103 | ||
| B. amyloliquefaciens LL3[ | Electroproration | Demethylation | pWH1520 | 7.6×102 |
| B. amyloliquefaciens[ | Chemical transformation | Overexpressing ComK regulator to induce the formation of competent | pUBXC | [(129 ± 20.6)- (1.7 ± 0.1)]×105 |
| PCR fragment-mediated knockout | [(3.2 ± 0.76)- (3.5 ± 0.42)]×104 | |||
| B. amyloliquefaciens[ | Transformation of protoplasts | Mediating by polyethylene glycol | pUB110 | (2-4)×105 |
Table 2 Study on the transformation efficiency of B. amyloliquefaciens by different methods
| Strain | Transformation method | Concrete operation | Transformation plasmid | Transformation efficiency /(CFU·μg-1) |
|---|---|---|---|---|
| B. amyloliquefaciens TA208[ | Electroproration | Adding DL-threonine or glycine to weaken the cell wall in hypertonic medium | pUB110 | (1.13 ± 0.34)×107 |
| Adding DL-threonine or glycine to weaken the cell wall in hypertonic medium;heat shock treatment | pHCMC02 | (8.94 ± 0.77)×105 | ||
| B. amyloliquefaciens NB[ | Electroproration | Demethylation | pDR | 4.94 ± 0.42)×104 |
| Demethylation | pMA5 | (6.15 ± 0.19)×103 | ||
| B. amyloliquefaciens LL3[ | Electroproration | Demethylation | pWH1520 | 7.6×102 |
| B. amyloliquefaciens[ | Chemical transformation | Overexpressing ComK regulator to induce the formation of competent | pUBXC | [(129 ± 20.6)- (1.7 ± 0.1)]×105 |
| PCR fragment-mediated knockout | [(3.2 ± 0.76)- (3.5 ± 0.42)]×104 | |||
| B. amyloliquefaciens[ | Transformation of protoplasts | Mediating by polyethylene glycol | pUB110 | (2-4)×105 |
| Products | Strategies | Yield | Reference | |
|---|---|---|---|---|
| γ-PGA | Knock out genes involved in by-products,degrading enzymes and optimize the endogenous glutamate synthesis | 20.3 g/L | [ | |
| Modification of substrate inulin utilization,sugar metabolism and by-product pathways | 32.14 g/L | [ | ||
| Using the CRISPRi system’s multiple sgRNA combination strategy to control the expression of degrading enzymes,that achieve multiple molecular weights of γ-PGA | Different molecular weights of γ-PGA High(>800 kD)、middle(400-600 kD)、low(50-100 kD);25-27 g/L | [ | ||
| To control the stereochemical configuration | Low-molecular-weight(<10 kD)γ-PGA;28.35 g/L | [ | ||
| EPS | Optimization of fermentation conditions | 4.46 g/L | [ | |
| Levan | Removing six protease-related genes,the biofilm matrix protein TasA and γ-PGA synthase genes | 31.1 g/L | [ | |
| Optimization of levansucrase expression based on promoters and signal peptides | 102 g/L | [ | ||
| Enzymes production | α-amylase | Optimization of the promoters and host | 2714 U/mL | [ |
| Pullulanase | Cloning and expression optimization | 2.8 ASPU/mL | [ | |
| Keratinase | Cloning and expression optimization | 1 361.54 U/mL | [ | |
| lytic polysaccharide monooxygenase | Establishing a high-throughput screening system for expression and secretion | 12.17 U/g | [ | |
| Nucleotides | Guanosine | Relieving the purine operon of the purine biosynthetic pathway and optimizing the energy of the respiratory chain | 19 g/L | [ |
| Inosine | Protoplast fusion | 6 g/L | [ | |
| Antimicrobial lipopeptides | Surfactin | Using nano iron particles to improve the permeability of cell membranes | 7.15 g/L | [ |
| Iturin A | Overexpression of the iturin A biosynthesis genes | 37.35 mg/L | [ | |
| Fengycin | Genome shuffling | 450.51 mg/L | [ | |
| Bacillomycin D | Knocking out of the gene rapC | (360.8±30.7)mg/L | [ | |
| Bulk chemicals | Acetoin | Compound mutagenesis | 85.2 g/L | [ |
| 2, 3-butanediol | Overexpression of glyceraldehyde-3-phosphate dehydrogenase and 2, 3-butanediol dehydrogenase | 132.9 g/L | [ | |
Table 3 Summary of the synthesis of biobased products based on B. amyloliquefaciens species
| Products | Strategies | Yield | Reference | |
|---|---|---|---|---|
| γ-PGA | Knock out genes involved in by-products,degrading enzymes and optimize the endogenous glutamate synthesis | 20.3 g/L | [ | |
| Modification of substrate inulin utilization,sugar metabolism and by-product pathways | 32.14 g/L | [ | ||
| Using the CRISPRi system’s multiple sgRNA combination strategy to control the expression of degrading enzymes,that achieve multiple molecular weights of γ-PGA | Different molecular weights of γ-PGA High(>800 kD)、middle(400-600 kD)、low(50-100 kD);25-27 g/L | [ | ||
| To control the stereochemical configuration | Low-molecular-weight(<10 kD)γ-PGA;28.35 g/L | [ | ||
| EPS | Optimization of fermentation conditions | 4.46 g/L | [ | |
| Levan | Removing six protease-related genes,the biofilm matrix protein TasA and γ-PGA synthase genes | 31.1 g/L | [ | |
| Optimization of levansucrase expression based on promoters and signal peptides | 102 g/L | [ | ||
| Enzymes production | α-amylase | Optimization of the promoters and host | 2714 U/mL | [ |
| Pullulanase | Cloning and expression optimization | 2.8 ASPU/mL | [ | |
| Keratinase | Cloning and expression optimization | 1 361.54 U/mL | [ | |
| lytic polysaccharide monooxygenase | Establishing a high-throughput screening system for expression and secretion | 12.17 U/g | [ | |
| Nucleotides | Guanosine | Relieving the purine operon of the purine biosynthetic pathway and optimizing the energy of the respiratory chain | 19 g/L | [ |
| Inosine | Protoplast fusion | 6 g/L | [ | |
| Antimicrobial lipopeptides | Surfactin | Using nano iron particles to improve the permeability of cell membranes | 7.15 g/L | [ |
| Iturin A | Overexpression of the iturin A biosynthesis genes | 37.35 mg/L | [ | |
| Fengycin | Genome shuffling | 450.51 mg/L | [ | |
| Bacillomycin D | Knocking out of the gene rapC | (360.8±30.7)mg/L | [ | |
| Bulk chemicals | Acetoin | Compound mutagenesis | 85.2 g/L | [ |
| 2, 3-butanediol | Overexpression of glyceraldehyde-3-phosphate dehydrogenase and 2, 3-butanediol dehydrogenase | 132.9 g/L | [ | |
| [1] | 张娟, 杨彩梅, 曹广添, 等. 解淀粉芽孢杆菌及其作为益生菌的应用[J]. 动物营养学报, 2014, 26(4):863-867. |
| Zhang J, Yang CM, Cao GT, et al. Bacillus amyloliquefaciens and its application as a probiotic[J]. Chin J Animal Nutr, 2014, 26(4):863-867. | |
| [2] |
Chen XT, Ji JB, Liu YC, et al. Artificial induction of genetic competence in Bacillus amyloliquefaciens isolates[J]. Biotechnol Lett, 2016, 38(12):2109-2117.
doi: 10.1007/s10529-016-2194-0 URL |
| [3] |
Qiu Y, Zhu Y, Zhang Y, et al. Characterization of a Regulator pgsR on Endogenous Plasmid p2Sip and Its Complementation for poly(γ-glutamic acid)accumulation in Bacillus amyloliquefaciens[J]. J Agric Food Chem, 2019, 67(13):3711-3722.
doi: 10.1021/acs.jafc.9b00332 URL |
| [4] |
Sha Y, Zhang Y, Qiu Y, et al. Efficient biosynjournal of low-molecular-weight poly-γ-glutamic acid by stable overexpression of PgdS hydrolase in Bacillus amyloliquefaciens NB[J]. J Agric Food Chem, 2019, 67(1):282-290.
doi: 10.1021/acs.jafc.8b05485 URL |
| [5] |
Zhang W, Xie H, He Y, et al. Chromosome integration of the Vitreoscilla hemoglobin gene(vgb)mediated by temperature-sensitive plasmid enhances γ-PGA production in Bacillus amyloliquefaciens[J]. FEMS Microbiol Lett, 2013, 343(2):127-134.
doi: 10.1111/1574-6968.12139 pmid: 23521121 |
| [6] | Zhou X, Zhang N, Xia LM, et al. ResDE two-component regulatory system mediates oxygen limitation-induced biofilm formation by Bacillus amyloliquefaciens SQR9[J]. Appl Environ Microbiol, 2018, 84(8):e02744-17. |
| [7] |
Guo X, Chai CC, An YJ, et al. Rational design of signal peptides for improved MtC1LPMO production in Bacillus amyloliquefaciens[J]. Int J Biol Macromol, 2021, 175:262-269.
doi: 10.1016/j.ijbiomac.2021.02.034 URL |
| [8] | 陈玉娟, 沈微, 陈献忠, 等. 解淀粉芽孢杆菌β-1, 3-1, 4-葡聚糖酶的高效表达[J]. 生物技术, 2011, 21(2):22-26. |
| Chen YJ, Shen W, Chen XZ, et al. Overexpression of Bacillus amyloliquefaciens β-1, 3-1, 4-glucanase[J]. Biotechnology, 2011, 21(2):22-26. | |
| [9] |
Gu Y, Zheng J, Feng J, et al. Improvement of levan production in Bacillus amyloliquefaciens through metabolic optimization of regulatory elements[J]. Appl Microbiol Biotechnol, 2017, 101(10):4163-4174.
doi: 10.1007/s00253-017-8171-2 URL |
| [10] |
Qiao JQ, Tian DW, Huo R, et al. Functional analysis and application of the cryptic plasmid pBSG3 harboring the RapQ-PhrQ system in Bacillus amyloliquefaciens B3[J]. Plasmid, 2011, 65(2):141-149.
doi: 10.1016/j.plasmid.2010.11.008 URL |
| [11] |
Liao Y, Huang L, Wang B, et al. The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data[J]. Gene, 2015, 571(2):252-262.
doi: 10.1016/j.gene.2015.06.066 URL |
| [12] |
Liao Y, Wang B, Ye Y, et al. Determination and optimization of a strong promoter element from Bacillus amyloliquefaciens by using a promoter probe vector[J]. Biotechnol Lett, 2018, 40(1):119-126.
doi: 10.1007/s10529-017-2449-4 URL |
| [13] |
Qiu YB, Zhu YF, Sha YY, et al. Development of a robust Bacillus amyloliquefaciens cell factory for efficient poly(γ-glutamic acid)production from Jerusalem artichoke[J]. ACS Sustainable Chem Eng, 2020, 8(26):9763-9774.
doi: 10.1021/acssuschemeng.0c02107 URL |
| [14] |
Novikov AA, Borukhov SI, Strongin AY. Bacillusamyloliquefaciens α -amylase signal sequence fused in frame with human proinsulin is properly processed by Bacillus subtilis cells[J]. Biochem Biophys Res Commun, 1990, 169(1):297-301.
doi: 10.1016/0006-291X(90)91467-7 URL |
| [15] |
Zakataeva NP, Nikitina OV, Gronskiy SV, et al. A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains[J]. Appl Microbiol Biotechnol, 2010, 85(4):1201-1209.
doi: 10.1007/s00253-009-2276-1 pmid: 19820923 |
| [16] | 杨慧林, 王坤, 廖瑜玲, 等. 解淀粉芽孢杆菌ptsGHI基因的敲除及缺陷株生长特性[J]. 华南理工大学学报:自然科学版, 2012, 40(8):95-100. |
| Yang HL, Wang K, Liao YL, et al. Knockout of ptsGHI gene of Bacillus amyloliquefaciens and growth characteristics of corresponding deficient strain[J]. J South China Univ Technol:Nat Sci Ed, 2012, 40(8):95-100. | |
| [17] |
Wu L, Wu H, Chen L, et al. Bacilysin overproduction in Bacillus amyloliquefaciens FZB42 markerless derivative strains FZBREP and FZBSPA enhances antibacterial activity[J]. Appl Microbiol Biotechnol, 2015, 99(10):4255-4263.
doi: 10.1007/s00253-014-6251-0 URL |
| [18] |
Zhang W, Gao WX, Feng J, et al. A markerless gene replacement method for B. amyloliquefaciens LL3 and its use in genome reduction and improvement of poly-γ-glutamic acid production[J]. Appl Microbiol Biotechnol, 2014, 98(21):8963-8973.
doi: 10.1007/s00253-014-5824-2 pmid: 24859524 |
| [19] |
Zhou C, Shi L, Ye B, et al. pheS *, an effective host-genotype-independent counter-selectable marker for marker-free chromosome deletion in Bacillus amyloliquefaciens[J]. Appl Microbiol Biotechnol, 2017, 101(1):217-227.
doi: 10.1007/s00253-016-7906-9 URL |
| [20] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6):1262-1278.
doi: 10.1016/j.cell.2014.05.010 URL |
| [21] |
Sha YY, Qiu YB, Zhu YF, et al. CRISPRi-based dynamic regulation of hydrolase for the synjournal of poly-γ-glutamic acid with variable molecular weights[J]. ACS Synth Biol, 2020, 9(9):2450-2459.
doi: 10.1021/acssynbio.0c00207 URL |
| [22] |
Zhang GQ, Bao P, Zhang Y, et al. Enhancing electro-transformation competency of recalcitrant Bacillus amyloliquefaciens by combining cell-wall weakening and cell-membrane fluidity disturbing[J]. Anal Biochem, 2011, 409(1):130-137.
doi: 10.1016/j.ab.2010.10.013 URL |
| [23] |
Vehmaanperä J. Transformation of Bacillus amyloliquefaciens protoplasts with plasmid DNA[J]. FEMS Microbiol Lett, 1988, 49(1):101-105.
doi: 10.1111/fml.1988.49.issue-1 URL |
| [24] |
Xue GP, Johnson JS, Dalrymple BP. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis[J]. J Microbiol Methods, 1999, 34(3):183-191.
doi: 10.1016/S0167-7012(98)00087-6 URL |
| [25] | Akamatsu T, Sekiguchi J. Transformation of Bacillus protoplasts by plasmid pTP4 DNA[J]. Agricultural and Biological Chemistry, 1982, 46(6):1617-1621. |
| [26] |
Wang P, Wang P, Tian J, et al. A new strategy to express the extracellular α-amylase from Pyrococcus furiosus in Bacillus amyloliquefaciens[J]. Sci Rep, 2016, 6:22229.
doi: 10.1038/srep22229 URL |
| [27] | 孙娟娟, 沈微, 石贵阳, 等. 普鲁兰酶基因在解淀粉芽孢杆菌BF7658菌株中的分泌表达[J]. 工业微生物, 2012, 42(2):18-22. |
| Sun JJ, Shen W, Shi GY, et al. Secreted expression of the pullulanase in Bacillus amyloliquefaciens BF7658[J]. Ind Microbiol, 2012, 42(2):18-22. | |
| [28] | 辛青龙, 刘业学, 何光明, 等. 地衣芽孢杆菌角蛋白酶kerA基因的异源表达及性能研究[J]. 中国皮革, 2018, 47(12):28-34. |
| Xin QL, Liu YX, He GM, et al. Overexpression of Bacillus licheniformis keratinase gene(kerA)and its performance[J]. China Leather, 2018, 47(12):28-34. | |
| [29] | 王慧. 解淀粉芽孢杆菌Bacillus amyloliquefaciens K11高效表达体系的建立及其高效表达元件的优化[D]. 北京:中国农业科学院, 2018. |
| Wang H. Establishment of the highly effective expression system in Bacillus amyloliquefaciens K11 and optimization of its efficient expression elements[D]. Beijing:Chinese Academy of Agricultural Sciences, 2018. | |
| [30] | 刘刚, 王清明, 陈惠鹏. 非经典的蛋白质分泌途径[J]. 生物技术通讯, 2005, 16(1):53-55. |
| Liu G, Wang QM, Chen HP. Non-classical secretory pathway[J]. Lett Biotechnol, 2005, 16(1):53-55. | |
| [31] | 徐虹, 冯小海, 徐得磊, 等. 聚氨基酸功能高分子的发展状况与应用前景[J]. 生物产业技术, 2017(6):92-99. |
| Xu H, Feng XH, Xu DL, et al. Recent developments and potential applications of poly(amino acid)-based functional polymers[J]. Biotechnol Bus, 2017(6):92-99. | |
| [32] |
Qiu Y, Sha Y, Zhang Y, et al. Development of Jerusalem artichoke resource for efficient one-step fermentation of poly-(γ-glutamic acid)using a novel strain Bacillus amyloliquefaciens NX-2S[J]. Bioresour Technol, 2017, 239:197-203.
doi: 10.1016/j.biortech.2017.05.005 URL |
| [33] |
Qiu YB, Zhang YT, Zhu YF, et al. Improving poly-(γ-glutamic acid)production from a glutamic acid-independent strain from inulin substrate by consolidated bioprocessing[J]. Bioprocess Biosyst Eng, 2019, 42(10):1711-1720.
doi: 10.1007/s00449-019-02167-w URL |
| [34] |
Sha Y, Huang Y, Zhu Y, et al. Efficient biosynjournal of low-molecular-weight poly-γ-glutamic acid based on stereochemistry regulation in Bacillus amyloliquefaciens[J]. ACS Synth Biol, 2020, 9(6):1395-1405.
doi: 10.1021/acssynbio.0c00080 URL |
| [35] |
Zhao W, Zhang J, Jiang YY, et al. Characterization and antioxidant activity of the exopolysaccharide produced by Bacillus amyloliquefaciens GSBa-1[J]. J Microbiol Biotechnol, 2018, 28(8):1282-1292.
doi: 10.4014/jmb.1801.01012 URL |
| [36] |
Han YZ, Liu EQ, Liu LS, et al. Rheological, emulsifying and thermostability properties of two exopolysaccharides produced by Bacillus amyloliquefaciens LPL061[J]. Carbohydr Polym, 2015, 115:230-237.
doi: 10.1016/j.carbpol.2014.08.044 URL |
| [37] |
Chen YT, Yuan Q, Shan LT, et al. Antitumor activity of bacterial exopolysaccharides from the endophyte Bacillus amyloliquefaciens sp. isolated from Ophiopogon japonicus.[J]. Oncol Lett, 2013, 5(6):1787-1792.
doi: 10.3892/ol.2013.1284 URL |
| [38] |
Yang H, Deng J, Yuan Y, et al. Two novel exopolysaccharides from Bacillus amyloliquefaciens C-1:antioxidation and effect on oxidative stress[J]. Curr Microbiol, 2015, 70(2):298-306.
doi: 10.1007/s00284-014-0717-2 URL |
| [39] |
Cai G, Liu Y, Li X, et al. New levan-type exopolysaccharide from Bacillus amyloliquefaciens as an antiadhesive agent against enterotoxigenic Escherichia coli[J]. J Agric Food Chem, 2019, 67(28):8029-8034.
doi: 10.1021/acs.jafc.9b03234 URL |
| [40] |
Feng J, Gu Y, Quan Y, et al. Recruiting a new strategy to improve levan production in Bacillus amyloliquefaciens[J]. Sci Rep, 2015, 5:13814.
doi: 10.1038/srep13814 pmid: 26347185 |
| [41] |
Phengnoi P, Charoenwongpaiboon T, Wangpaiboon K, et al. Levansucrase from Bacillus amyloliquefaciens KK9 and its Y237S variant producing the high bioactive levan-type fructooligosaccharides[J]. Biomolecules, 2020, 10(5):692.
doi: 10.3390/biom10050692 URL |
| [42] | 吴飞, 史建明, 谢希贤, 等. 鸟苷产生菌解淀粉芽孢杆菌的选育[J]. 天津科技大学学报, 2010, 25(2):1-4. |
| Wu F, Shi JM, Xie XX, et al. Breeding of strain Bacillus amyloliquefaciens for guanosine-producing[J]. J Tianjin Univ Sci Technol, 2010, 25(2):1-4. | |
| [43] | 何逵夫, 马跃超, 杜姗姗, 等. 解淀粉芽胞杆菌关键酶基因过表达对鸟苷积累的影响[J]. 微生物学报, 2012, 52(6):718-725. |
| He KF, Ma YC, Du SS, et al. Effects of overexpression of key enzyme genes on guanosine accumulation in Bacillus amyloliquefaciens[J]. Acta Microbiol Sin, 2012, 52(6):718-725. | |
| [44] |
Liao YL, Ye YR, Wang B, et al. Optimization of the purine operon and energy generation in Bacillus amyloliquefaciens for guanosine production[J]. Biotechnol Lett, 2017, 39(11):1675-1682.
doi: 10.1007/s10529-017-2412-4 URL |
| [45] |
Wong JH, Hao J, Cao Z, et al. An antifungal protein from Bacillus amyloliquefaciens[J]. J Appl Microbiol, 2008, 105(6):1888-1898.
doi: 10.1111/j.1365-2672.2008.03917.x pmid: 19120637 |
| [46] | 吴江, 吴梧桐, 黄为一, 等. 通过原生质体融合选育直接利用淀粉的肌苷产生菌[J]. 药物生物技术, 1995, 2(2):7-10 |
| Wu J, Wu WT, Huang WY, et al. Selection of inosine producing fusants by protoplast fusion[J]. Pharm Biotechnol, 1995, 2(2):7-10 | |
| [47] |
Wibisana A, Sumaryono W, Mirawati Sudiro T, et al. Optimization of surfactin production by Bacillus amyloliquefaciens MD4-12 using response surface methodology[J]. Microbiol Indones, 2015, 9(3):120-128.
doi: 10.5454/mi.9.3.4 URL |
| [48] | 周泽宇, 张婉茹, 张柔萱, 等. 代谢工程改造Bacillus amyloliquefaciens提高Surfactin产量[J]. 南开大学学报:自然科学版, 2018, 51(5):18-26. |
| Zhou ZY, Zhang WR, Zhang RX, et al. Metabolic engineering of Bacillus amyloliquefaciens to improve surfactin production[J]. Acta Sci Nat Univ Nankaiensis, 2018, 51(5):18-26. | |
| [49] |
Yang N, Wu Q, Xu Y. Fe nanoparticles enhanced surfactin production in Bacillus amyloliquefaciens[J]. ACS Omega, 2020, 5(12):6321-6329.
doi: 10.1021/acsomega.9b03648 pmid: 32258866 |
| [50] |
Dang Y, Zhao F, Liu X, et al. Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization[J]. Microb Cell Fact, 2019, 18(1):68.
doi: 10.1186/s12934-019-1121-1 URL |
| [51] |
Sang-Cheol L, Kim SH, Park IH, et al. Isolation, purification, and characterization of novel fengycin S from Bacillus amyloliquefaciens LSC04 degrading-crude oil[J]. Biotechnol Bioprocess Eng, 2010, 15(2):246-253.
doi: 10.1007/s12257-009-0037-8 URL |
| [52] |
Zhao JF, Zhang C, Lu J, et al. Enhancement of fengycin production in Bacillus amyloliquefaciens by genome shuffling and relative gene expression analysis using RT-PCR[J]. Can J Microbiol, 2016, 62(5):431-436.
doi: 10.1139/cjm-2015-0734 URL |
| [53] | Sun HG, Lu FX, Zhang C, et al. Improvement of fengycin production by Bacillus amyloliquefaciens via promoter replacement at the fengycin operon with the P59 and PrepU promoters[J]. Journal of Pure and Applied Microbiology, 2014, 8(2):1071-1077. |
| [54] | Sun J, Liu Y, Lin F, et al. CodY, ComA, DegU and Spo0A controlling lipopeptides biosynjournal in Bacillus amyloliquefaciens fmb[J][J]. J Appl Microbiol, 2021:1364-5072. |
| [55] |
Zhang YJ, Li SB, Liu LM, et al. Acetoin production enhanced by manipulating carbon flux in a newly isolated Bacillus amyloliquefaciens[J]. Bioresour Technol, 2013, 130:256-260.
doi: 10.1016/j.biortech.2012.10.036 URL |
| [56] |
Luo QL, Wu J, Wu MC. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance[J]. Process Biochem, 2014, 49(8):1223-1230.
doi: 10.1016/j.procbio.2014.05.005 URL |
| [57] | 王诗卉, 罗秋玲, 刘佳, 等. 高产3-羟基丁酮解淀粉芽孢杆菌的选育及发酵优化[J]. 生物工程学报, 2018, 34(5):803-811. |
| Wang SH, Luo QL, Liu J, et al. Mutation and fermentation optimization of Bacillus amyloliquefaciens for acetoin production[J]. Chin J Biotechnol, 2018, 34(5):803-811. | |
| [58] |
Yang TW, Rao ZM, Zhang X, et al. Production of 2, 3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens[J]. J Basic Microbiol, 2011, 51(6):650-658.
doi: 10.1002/jobm.v51.6 URL |
| [59] |
Yang T, Rao Z, Zhang X, et al. Improved production of 2, 3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2, 3-butanediol dehydrogenase[J]. PLoS One, 2013, 8(10):e76149.
doi: 10.1371/journal.pone.0076149 URL |
| [60] |
Feng J, Gu YY, Quan YF, et al. Improved poly-γ-glutamic acid production in Bacillus amyloliquefaciens by modular pathway engineering[J]. Metab Eng, 2015, 32:106-115.
doi: S1096-7176(15)00121-4 pmid: 26410449 |
| [61] | 李彦岩, 张彩, 范熠, 等. 一株解淀粉芽孢杆菌产糖条件的优化[J]. 食品科学, 2013, 34(7):185-189. |
| Li YY, Zhang C, Fan Y, et al. Optimization of fermentation conditions for exopolysaccharide production by Bacillus amyloliquefaciens LPL061[J]. Food Sci, 2013, 34(7):185-189. | |
| [62] |
Sun J, Qian S, Lu J, et al. Knockout of rapC improves the bacillomycin D yield based on de novo genome sequencing of Bacillus amyloliquefaciens fmb[J]. J Agric Food Chem, 2018, 66(17):4422-4430.
doi: 10.1021/acs.jafc.8b00418 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [3] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [4] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [5] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [6] | YU Hui-li, LI Ai-tao. Application of Cytochrome P450 in the Biosynthesis of Flavors and Fragrances [J]. Biotechnology Bulletin, 2023, 39(4): 24-37. |
| [7] | JIANG Jing-jing, ZHOU Zhao-xu, DU Hui, LYU Zhao-long, WANG Chun-ming, GUO Jian-guo, ZHANG Xin-rui, LI Ji-ping. Isolation and Identification of Apple Brown Rot Pathogen in Parts of Gansu and Screening of Antagonistic Bacteria [J]. Biotechnology Bulletin, 2023, 39(10): 209-218. |
| [8] | NIU Xin, ZHANG Ying, WANG Mao-jun, LIU Wen-long, LU Fu-ping, LI Yu. Effects of Different Integration Sites on the Expression of Exogenous Alkaline Protease in Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2022, 38(4): 253-260. |
| [9] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| [10] | YANG Rui-xian, LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da. Analysis of Antimicrobial Active Metabolites from Antagonistic Strains Against Fusarium solani [J]. Biotechnology Bulletin, 2022, 38(2): 57-66. |
| [11] | CAI Guo-lei, LU Xiao-kai, LOU Shui-zhu, YANG Hai-ying, DU Gang. Classification and Identification of Bacillus LM Based on Whole Genome and Study on Its Antibacterial Principle [J]. Biotechnology Bulletin, 2021, 37(8): 176-185. |
| [12] | LI Jin, PENG Ke-wei, PAN Qiu-yi, ZHU Zhe-yuan, PENG Di. Isolation and Identification of Bacillus amyloliquefaciens HR-2 and Biological Control of Rice Blast [J]. Biotechnology Bulletin, 2021, 37(3): 27-34. |
| [13] | LI Xin-yue, ZHANG Jin-fang, XU Xiao-jian, LU Fu-ping, LI Yu. Effects of Spore Formation Related Gene Deletion on Biomass and Extracellular Enzyme Expression of Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2021, 37(3): 35-43. |
| [14] | YE Jian-wen, CHEN Jiang-nan, ZHANG Xu, Wu Fu-qing, CHEN Guo-qiang. Dynamic Control:An Efficient Strategy for Metabolically Engineering Microbial Cell Factories [J]. Biotechnology Bulletin, 2020, 36(6): 1-12. |
| [15] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||