








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 173-180.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0647
Previous Articles Next Articles
YAN Jiong1( ), FENG Chen-yi1, GAO Xue-kun1, XU Xiang1, YANG Jia-min1, CHEN Zhao-yang2(
), FENG Chen-yi1, GAO Xue-kun1, XU Xiang1, YANG Jia-min1, CHEN Zhao-yang2( )
)
Received:2021-05-19
Online:2022-03-26
Published:2022-04-06
Contact:
CHEN Zhao-yang
E-mail:yanjiong@126.com;ccytycn@163.com
YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology[J]. Biotechnology Bulletin, 2022, 38(3): 173-180.
| 名称 Name | 序列 Sequence(5'-3') | PAM序列 PAM sequence |
|---|---|---|
| sgRNA-Plin1-1 | TTGGAGGGCGAATGTTACAT | AGG |
| sgRNA-Plin1-2 | GAACTTCAGGCGTATGACCC | TGG |
Table 1 sgRNA sequence
| 名称 Name | 序列 Sequence(5'-3') | PAM序列 PAM sequence |
|---|---|---|
| sgRNA-Plin1-1 | TTGGAGGGCGAATGTTACAT | AGG |
| sgRNA-Plin1-2 | GAACTTCAGGCGTATGACCC | TGG |
| 名称Name | 序列Sequence(5'-3') | 大小Size/bp |
|---|---|---|
| Plin1-F | CTGAGAGAAGGCTTAACCTTGCTGG | 25 |
| Plin1-R1 | AGCTTTCCATCCTGCAAGTGAGTCAG | 26 |
| Plin1-R2 | AGGTGGCAAGGACAGAGACAGTGAG | 25 |
Table 2 Primer sequence for genotyping
| 名称Name | 序列Sequence(5'-3') | 大小Size/bp |
|---|---|---|
| Plin1-F | CTGAGAGAAGGCTTAACCTTGCTGG | 25 |
| Plin1-R1 | AGCTTTCCATCCTGCAAGTGAGTCAG | 26 |
| Plin1-R2 | AGGTGGCAAGGACAGAGACAGTGAG | 25 |
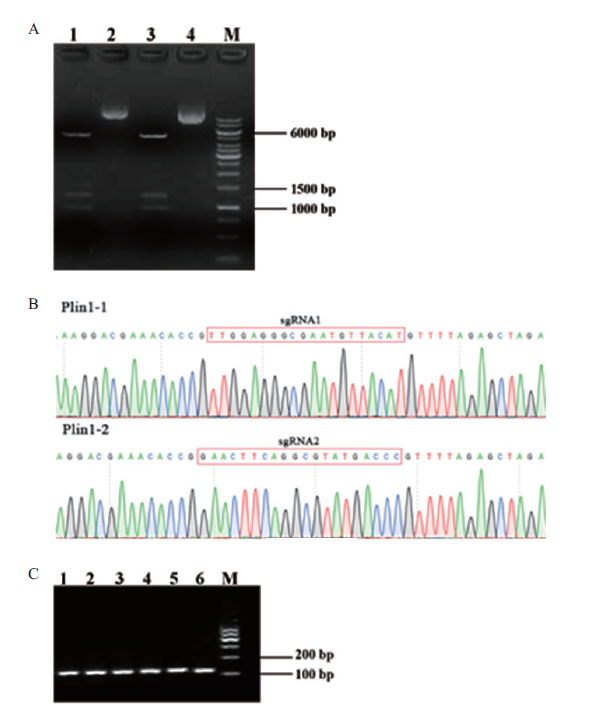
Fig. 3 Construction and identification of sgRNA expression vector A:Digestion results of sgRNA expression vector(Lane 2 and 4:sgRNA-Plin1-1,2 vectors. Lane 1 and 3:Enzyme fragment. M:DNA marker). B:Sequencing results of sgRNA expression vector. C:In vitro transcriptional fragment electrophoresis results of sgRNA(Lane 1-3:sgRNA-Plin1-1 transcription products. Lane 4-6 lane:sgRNA-Plin1-2 transcription products. M:DNA marker)
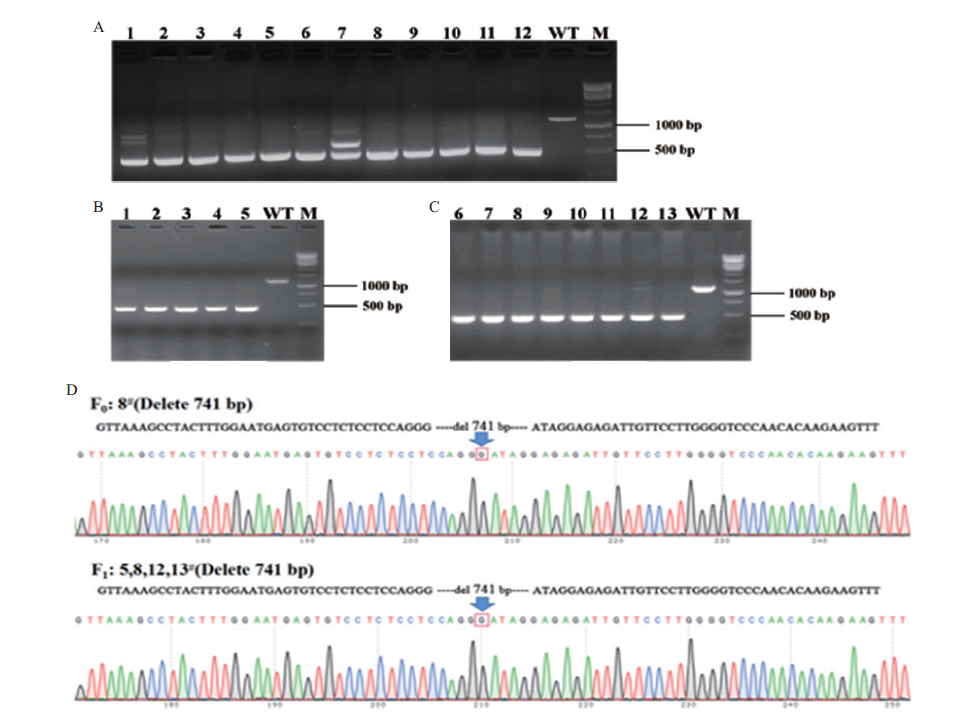
Fig. 4 Genotype identification of F0 and F1 generation mice A:Electrophoretic results of Plin1-F1/R1 amplified from F0 generation positive mice;lane 1-12:F0 positive mice;WT:wild type control;M. DNA marker. B-C:Electrophoretic results of Plin1-F1/R1 amplified from F1 generation mice;lane 1-13:F1 generation mice;WT:wild type control. M:DNA marker. D:gene sequencing results of F0 and F1 generation mice
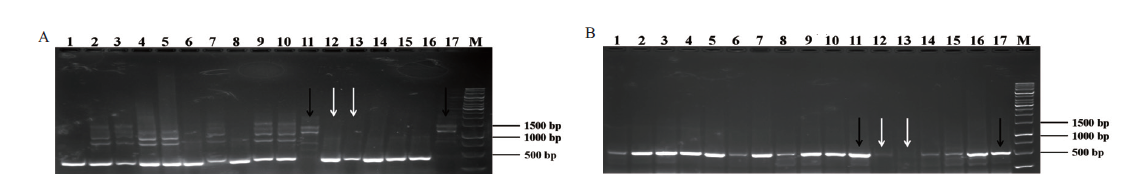
Fig. 5 Genotype identification of F2 generation mice A:Electrophoretic results of Plin1 gene amplified by Plin1-F1/R1. B:Electrophoretic results of Plin1 gene amplified by Plin1-F1/R2. Lane 1-17:Plin1 gene fragment of F2 generation mice. White arrow:Homozygous. Black arrow:Wild type
| 组别/指标 Group/index | 体重 Body weight/g | 体长 Body length/cm | 摄食量 Food intake/g |
|---|---|---|---|
| 野生型WT | 22.95±0.55 | 8.88±2.17 | 16.26±2.13 |
| 杂合型Plin1+/- | 23.20±1.57 | 9.31±3.49 | 15.43±3.03 |
| 纯合型Plin1-/- | 18.34±0.35* | 8.72±1.34 | 15.67±2.59 |
Table 3 Physical parameters of mice in each group
| 组别/指标 Group/index | 体重 Body weight/g | 体长 Body length/cm | 摄食量 Food intake/g |
|---|---|---|---|
| 野生型WT | 22.95±0.55 | 8.88±2.17 | 16.26±2.13 |
| 杂合型Plin1+/- | 23.20±1.57 | 9.31±3.49 | 15.43±3.03 |
| 纯合型Plin1-/- | 18.34±0.35* | 8.72±1.34 | 15.67±2.59 |
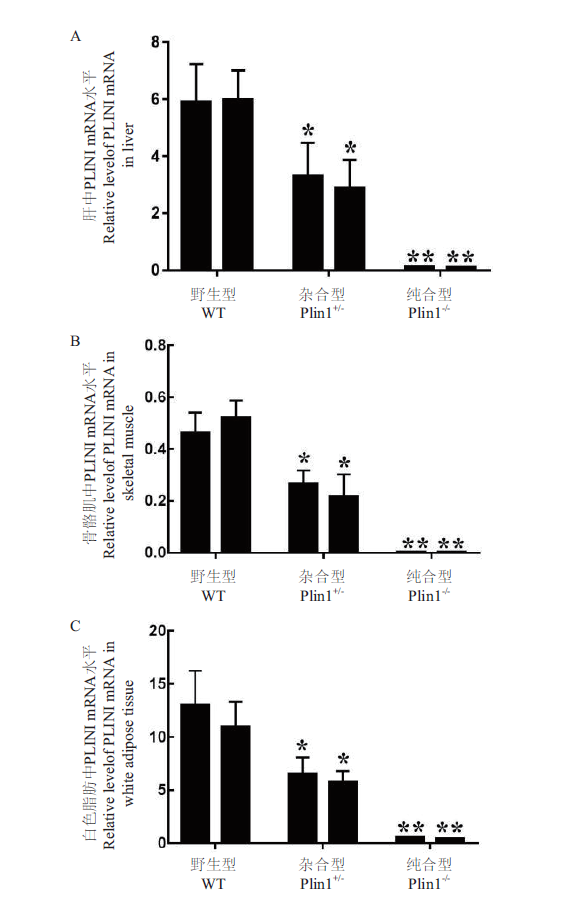
Fig. 6 Relative expressions of PLIN1 mRNA in the various tissues of mice A:Relative level of PLINI mRNA in liver. B:Relative level of PLINI mRNA in skeletal muscle. C:Relative level of PLINI mRNA in white adipose tissue. **:Com-pared with Plin1+/-. P < 0.05. *:Compared with WT,P < 0.05. The same below
| [1] |
Jarc E, Petan T. Lipid droplets and the management of cellular stress[J]. Yale J Biol Med, 2019, 92(3):435-452.
pmid: 31543707 |
| [2] |
Jackson CL. Lipid droplet biogenesis[J]. Curr Opin Cell Biol, 2019, 59:88-96.
doi: 10.1016/j.ceb.2019.03.018 URL |
| [3] |
Itabe H, Yamaguchi T, Nimura S, et al. Perilipins:a diversity of intracellular lipid droplet proteins[J]. Lipids Health Dis, 2017, 16(1):83.
doi: 10.1186/s12944-017-0473-y URL |
| [4] |
Huang X, Sun J, Bian C, et al. Perilipin 1-3 in grass carp Ctenopharyngodon idella:molecular characterization, gene structure, tissue distribution, and mRNA expression in DHA-induced lipid droplet formation in adipocytes[J]. Fish Physiol Biochem, 2020, 46(6):2311-2322.
doi: 10.1007/s10695-020-00857-x URL |
| [5] |
Westhoff CC, Mrozinski J, Riedel I, et al. Perilipin 1 is a highly specific marker for adipocytic differentiation in sarcomas with intermediate sensitivity[J]. J Cancer Res Clin Oncol, 2017, 143(2):225-232.
doi: 10.1007/s00432-016-2263-8 URL |
| [6] |
Zhang S, Liu G, Xu C, et al. Perilipin 1 mediates lipid metabolism homeostasis and inhibits inflammatory cytokine synjournal in bovine adipocytes[J]. Front Immunol, 2018, 9:467.
doi: 10.3389/fimmu.2018.00467 URL |
| [7] |
Sohn JH, Lee YK, Han JS, et al. Perilipin 1(Plin1)deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation[J]. J Biol Chem, 2018, 293(36):13974-13988.
doi: 10.1074/jbc.RA118.003541 URL |
| [8] |
de Oliveira BAP, de Souza Pinhel MA, Nicoletti CF, et al. UCP2 and PLIN1 expression affects the resting metabolic rate and weight loss on obese patients[J]. Obes Surg, 2017, 27(2):343-348.
doi: 10.1007/s11695-016-2275-0 pmid: 27376365 |
| [9] | Zou L, Wang W, Liu S, et al. Spontaneous hypertension occurs with adipose tissue dysfunction in perilipin-1 null mice[J]. Biochim Biophys Acta, 2016, 1862(2):182-191. |
| [10] |
Mori Y, Otabe S, Dina C, et al. Genome-wide search for type 2 diabetes in Japanese affected sib-pairs confirms susceptibility genes on 3q, 15q, and 20q and identifies two new candidate Loci on 7p and 11p[J]. Diabetes, 2002, 51(4):1247-1255.
doi: 10.2337/diabetes.51.4.1247 URL |
| [11] | 陈燕波, 常翠青, 黄志卓, 等. PLIN基因多态性在中国汉族成年肥胖者中的分布[J]. 营养学报, 2011, 33(1):29-33. |
| Chen YB, Chang CQ, Huang ZZ, et al. Distribution of plin gene polymorphism in Chinese Han obese adults[J]. Acta Nutr Sin, 2011, 33(1):29-33. | |
| [12] | Hryhorowicz M, Lipiński D, Zeyland J, et al. CRISPR/Cas9 immune system as a tool for genome engineering[J]. Arch Immunol Ther Exp:Warsz, 2017, 65(3):233-240. |
| [13] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121):819-823.
doi: 10.1126/science.1231143 pmid: 23287718 |
| [14] |
Huang J, Wang Y, Zhao J. CRISPR editing in biological and biomedical investigation[J]. J Cell Physiol, 2018, 233(5):3875-3891.
doi: 10.1002/jcp.v233.5 URL |
| [15] |
Mashiko D, Young SA, Muto M, et al. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes[J]. Dev Growth Differ, 2014, 56(1):122-129.
doi: 10.1111/dgd.2014.56.issue-1 URL |
| [16] |
许祥, 董维鹏, 张少华, 等. 围脂滴蛋白基因CRISPR/Cas9载体的活性分析[J]. 生物技术通报, 2019, 35(11):89-95.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0357 |
| Xu X, Dong WP, Zhang SH, et al. Construction and activity analysis of the PLIN1 gene CRISPR/Cas9 vector[J]. Biotechnol Bull, 2019, 35(11):89-95. | |
| [17] | Morales PE, Bucarey JL, Espinosa A. Muscle lipid metabolism:role of lipid droplets and perilipins[J]. J Diabetes Res, 2017, 2017:1789395. |
| [18] | Sztalryd C, Brasaemle DL. The perilipin family of lipid droplet proteins:Gatekeepers of intracellular lipolysis[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2017, 1862(10 pt b):1221-1232. |
| [19] |
Maurizi G, Petäistö T, Maurizi A, et al. Key-genes regulating the liposecretion process of mature adipocytes[J]. J Cell Physiol, 2018, 233(5):3784-3793.
doi: 10.1002/jcp.26188 pmid: 28926092 |
| [20] | 赵志武, 王君实, 马敏, 等. 下调perilipin 1基因表达对3T3-L1细胞脂解的影响[J]. 中国生物工程杂志, 2016, 36(3):17-22. |
| Zhao ZW, Wang JS, Ma M, et al. Effect of down-regulated perilipin 1 gene expression on lipolysis of 3T3-L1 adipocytes[J]. China Biotechnol, 2016, 36(3):17-22. | |
| [21] | 张少华, 赵志武, 王君实, 等. 沉默PLIN1基因与异丙肾上腺素对3T3-L1脂肪细胞脂解的机制探究[J]. 现代预防医学, 2018, 45(11):2023-2027, 2038. |
| Zhang SH, Zhao ZW, Wang JS, et al. Combined effect of PLIN1 gene silencing and isoproterenol on lipolysis of 3T3-L1 adipocytes[J]. Mod Prev Med, 2018, 45(11):2023-2027, 2038. | |
| [22] |
Tansey JT, Sztalryd C, Gruia-Gray J, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity[J]. PNAS, 2001, 98(11):6494-6499.
doi: 10.1073/pnas.101042998 pmid: 11371650 |
| [23] |
Miyoshi H, Souza SC, Endo M, et al. Perilipin overexpression in mice protects against diet-induced obesity[J]. J Lipid Res, 2010, 51(5):975-982.
doi: 10.1194/jlr.M002352 URL |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [6] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [7] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| [14] | ZONG Mei, HAN Shuo, GUO Ning, DUAN Meng-meng, LIU Fan, WANG Gui-xiang. Production of Marker-free Mutants of Brassica campestris Mediated by CRISPR/Cas9 Through Vacuum Infiltration [J]. Biotechnology Bulletin, 2022, 38(10): 159-163. |
| [15] | WANG Hai-jie, WANG Cheng-ji, GUO Yang, WANG Yun, CHEN Yan-juan, LIANG Min, WANG Jue, GONG Hui, SHEN Ru-ling. Construction of Coagulation Factor 8 Gene Knockout Mouse Model Based on CRSIPR/Cas9 Technique and Verification of Phenotype [J]. Biotechnology Bulletin, 2022, 38(10): 273-280. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||