








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (3): 234-245.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0341
Previous Articles Next Articles
SUN Man-luan1( ), GE Sai2, BU Jia1, ZHU Zhuang-yan1
), GE Sai2, BU Jia1, ZHU Zhuang-yan1
Received:2021-03-18
Online:2022-03-26
Published:2022-04-06
Contact:
SUN Man-luan
E-mail:dtusml@sxdtdx.edu.cn
SUN Man-luan, GE Sai, BU Jia, ZHU Zhuang-yan. Regulation Mechanism of Ribonucleases in Escherichia coli[J]. Biotechnology Bulletin, 2022, 38(3): 234-245.

Fig.1 RNase-dependent RNA processing and degradation in Escherichia coli A:RNA processing mediated by endoribonucleas/exoribonuclease. B:sRNA-mediated mRNA degradation. C:RNA degradation mediated by endoribonuclease/exoribonuclease
| 名称 Name | 编码基因 Gene | 酶活特异性 Enzyme activity | 主要生理功能 Physiological functions | 已知主要调控方式 Regulation known | 主要参考文献 Reference | |
|---|---|---|---|---|---|---|
| 核 糖 核 酸 内 切 酶 | RNAse I | rna | 内切酶活性,Ca2+依赖的双链mRNA活性 | 尚不明确 | 可能与周质空间及胞内定位调节有关 | [ |
| RNAse III | rnc | 双链特异性内切酶活性 | rRNA的成熟 | 转录后自我调控;酶蛋白磷酸化修饰;反式作用因子调控 | [ | |
| RNase BN | rbn | 内切酶/外切酶活性 | sRNA的降解,mRNA降解 | 自身mRNA稳定性调节 | [ | |
| RNase E | rne | 内切酶活性 | mRNA的降解;rRNA及tRNA的成熟 | 转录后自我调控;酶蛋白磷酸化修饰;反式作用因子调控;sRNA;小分子结合及胞内定位 | [ | |
| RNase P | rnpA/rnpB | 内切酶活性 | 5' tRNA的成熟 | 未知 | [ | |
| RNase G | rng | 内切酶活性 | rRNA的成熟 | 未知 | [ | |
| RNase H家族 | rnhA/rnhB | RNA/DNA杂交体中RNA的切割与修饰 | 参与DNA复制过程中RNA的调整 | 未知 | [ | |
| YbeY | ybeY | 内切酶活性 | rRNA成熟与质控,16S rRNA3'末端的加工 | 未知 | [ | |
| RNase LS | rnlA/rnlB | 内切酶活性 | 降解T4噬菌体编码的mRNA | 未知 | [ | |
| 核 糖 核 酸 外 切 酶 | PNPase | pnp | 磷酸盐依赖的3'-5'外切酶活性 | mRNA的降解;rRNA及tRNA的成熟;sRNAs调控伴侣 | 自我调控;反式作用因子调控;sRNA的调控 | [ |
| RNAse II | rnb | 3'-5'外切酶活性 | mRNA的降解;rRNA及tRNA的成熟 | 酶蛋白的乙酰化修饰 | [ | |
| RNase R | rnr | 3'-5'外切酶活性 | mRNA的降解;rRNA的成熟与质控;tRNA的成熟 | 酶蛋白的乙酰化修饰 | [ | |
| RNase PH | rph | 磷酸盐依赖的3'-5'外切酶活性 | tRNA与rRNA的成熟;rRNA的降解 | 蛋白稳定性调节;RNase II调节(待发表资料) | [ | |
| RNase T | rnt | 3'-5'外切酶活性 | tRNA与rRNA的成熟 | 未知 | [ | |
| RNase D | rnd | 3'-5'外切酶活性 | tRNA的成熟 | 未知 | [ | |
| Oligo RNase | orn | 3'-5'外切酶活性 | 短链核苷酸的去除 | 未知 | [ | |
| RNase AM | trpH/yciV | 5'-3'外切酶活性 | 5S、23S 及16S rRNA的5'末端成熟 | 未知 | [ | |
Table 1 Properties and regulations of RNases in Escherichia coli
| 名称 Name | 编码基因 Gene | 酶活特异性 Enzyme activity | 主要生理功能 Physiological functions | 已知主要调控方式 Regulation known | 主要参考文献 Reference | |
|---|---|---|---|---|---|---|
| 核 糖 核 酸 内 切 酶 | RNAse I | rna | 内切酶活性,Ca2+依赖的双链mRNA活性 | 尚不明确 | 可能与周质空间及胞内定位调节有关 | [ |
| RNAse III | rnc | 双链特异性内切酶活性 | rRNA的成熟 | 转录后自我调控;酶蛋白磷酸化修饰;反式作用因子调控 | [ | |
| RNase BN | rbn | 内切酶/外切酶活性 | sRNA的降解,mRNA降解 | 自身mRNA稳定性调节 | [ | |
| RNase E | rne | 内切酶活性 | mRNA的降解;rRNA及tRNA的成熟 | 转录后自我调控;酶蛋白磷酸化修饰;反式作用因子调控;sRNA;小分子结合及胞内定位 | [ | |
| RNase P | rnpA/rnpB | 内切酶活性 | 5' tRNA的成熟 | 未知 | [ | |
| RNase G | rng | 内切酶活性 | rRNA的成熟 | 未知 | [ | |
| RNase H家族 | rnhA/rnhB | RNA/DNA杂交体中RNA的切割与修饰 | 参与DNA复制过程中RNA的调整 | 未知 | [ | |
| YbeY | ybeY | 内切酶活性 | rRNA成熟与质控,16S rRNA3'末端的加工 | 未知 | [ | |
| RNase LS | rnlA/rnlB | 内切酶活性 | 降解T4噬菌体编码的mRNA | 未知 | [ | |
| 核 糖 核 酸 外 切 酶 | PNPase | pnp | 磷酸盐依赖的3'-5'外切酶活性 | mRNA的降解;rRNA及tRNA的成熟;sRNAs调控伴侣 | 自我调控;反式作用因子调控;sRNA的调控 | [ |
| RNAse II | rnb | 3'-5'外切酶活性 | mRNA的降解;rRNA及tRNA的成熟 | 酶蛋白的乙酰化修饰 | [ | |
| RNase R | rnr | 3'-5'外切酶活性 | mRNA的降解;rRNA的成熟与质控;tRNA的成熟 | 酶蛋白的乙酰化修饰 | [ | |
| RNase PH | rph | 磷酸盐依赖的3'-5'外切酶活性 | tRNA与rRNA的成熟;rRNA的降解 | 蛋白稳定性调节;RNase II调节(待发表资料) | [ | |
| RNase T | rnt | 3'-5'外切酶活性 | tRNA与rRNA的成熟 | 未知 | [ | |
| RNase D | rnd | 3'-5'外切酶活性 | tRNA的成熟 | 未知 | [ | |
| Oligo RNase | orn | 3'-5'外切酶活性 | 短链核苷酸的去除 | 未知 | [ | |
| RNase AM | trpH/yciV | 5'-3'外切酶活性 | 5S、23S 及16S rRNA的5'末端成熟 | 未知 | [ | |

Fig. 2 Regulation mechanisms of RNase E in E. coli A:Autoregulation of RNase E. B:Regulation of RNase E by post-translational modification(phosphorylation). C:sRNA-Hfq regulation. D:RNase E regulated by regulator proteins. E:RNase E regulated by localization. F:Other regulations(by small molecules,enzymes,and antibiotics)
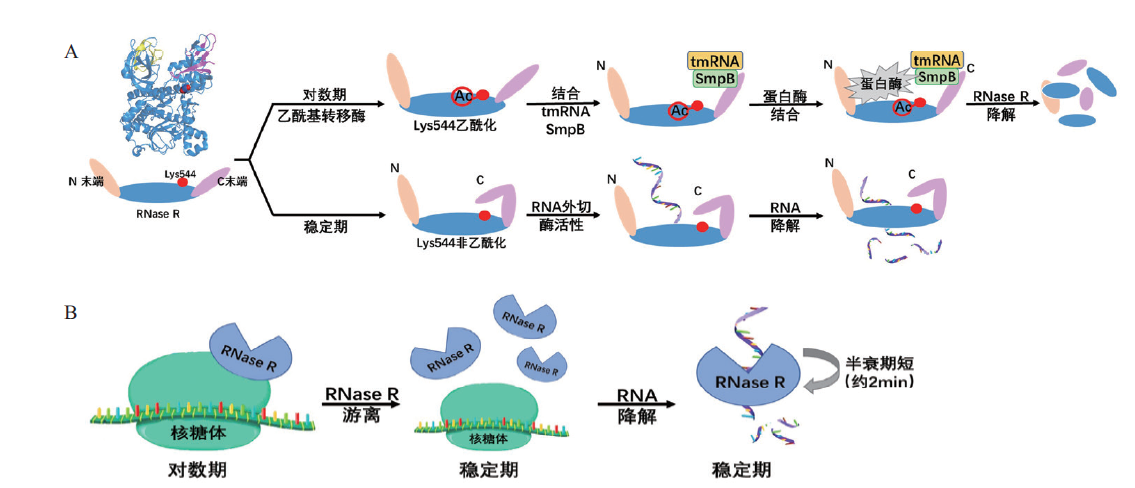
Fig. 4 Regulation mechanisms of RNase R in E. coli A:RNase R stability affected by post-translational modification(acetylation). B:Ribosome binding forms affect the activity and stability of RNase R
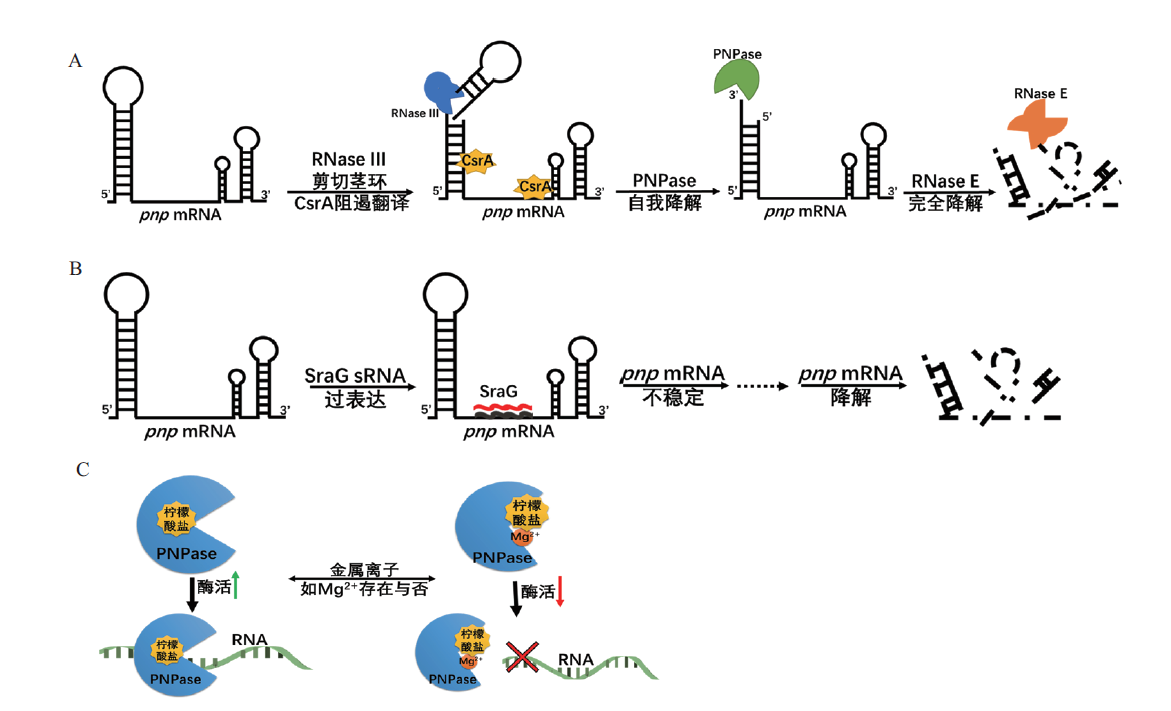
Fig. 5 Regulation mechanisms of PNPase in E. coli A:Autoregulation of PNPase. B:sRNA SraG mediated regulation of PNPase. C:Activity of PNPase modulated by citrate
| [1] |
Deutscher MP. Twenty years of bacterial RNases and RNA processing:how we’ve matured[J]. RNA, 2015, 21(4):597-600.
doi: 10.1261/rna.049692.115 pmid: 25780155 |
| [2] | de Trinquier A, Durand S, Braun F, et al. Regulation of RNA processing and degradation in bacteria[J]. BBA Gene Regul Mech, 2020, 1863(5):194505. |
| [3] |
Canestrari E, Paroo Z. Ribonucleases as drug targets[J]. Trends Pharmacol Sci, 2018, 39(10):855-866.
doi: S0165-6147(18)30121-4 pmid: 30144949 |
| [4] |
Vargas-Blanco DA, Shell SS. Regulation of mRNA stability during bacterial stress responses[J]. Front Microbiol, 2020, 11:2111.
doi: 10.3389/fmicb.2020.02111 pmid: 33013770 |
| [5] |
Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819):1709-1712.
doi: 10.1126/science.1138140 pmid: 17379808 |
| [6] | Roux C, Etienne TA, Hajnsdorf E, et al. The essential role of mRNA degradation in understanding and engineering E. coli metabolism[J]. Biotechnol Adv, 2021: 107805. |
| [7] | Stenum TS, Holmqvist E. CsrA enters Hfq's territory:Regulation of a base-pairing small RNA[J]. Mol Microbiol, 2021:mmi. 14785. |
| [8] |
Bechhofer DH, Deutscher MP. Bacterial ribonucleases and their roles in RNA metabolism[J]. Crit Rev Biochem Mol Biol, 2019, 54(3):242-300.
doi: 10.1080/10409238.2019.1651816 URL |
| [9] |
Zhu LQ, Gangopadhyay T, Padmanabha KP, et al. Escherichia coli RNA gene encoding RNase I:cloning, overexpression, subcellular distribution of the enzyme, and use of an RNA deletion to identify additional RNases[J]. J Bacteriol, 1990, 172(6):3146-3151.
pmid: 2188952 |
| [10] |
Altuvia Y, Bar A, Reiss N, et al. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli[J]. Nucleic Acids Res, 2018, 46(19):10380-10394.
doi: 10.1093/nar/gky684 pmid: 30113670 |
| [11] |
Lee M, Joo M, Sim M, et al. The coordinated action of RNase III and RNase G controls enolase expression in response to oxygen availability in Escherichia coli[J]. Sci Rep, 2019, 9(1):17257.
doi: 10.1038/s41598-019-53883-y URL |
| [12] | Altuvia S, Storz G, Papenfort K. Cross-regulation between bacteria and phages at a posttranscriptional level[J]. Microbiol Spectr, 2018, 6(4):10. 1128. |
| [13] |
Chu LY, Hsieh TJ, Golzarroshan B, et al. Structural insights into RNA unwinding and degradation by RNase R[J]. Nucleic Acids Res, 2017, 45(20):12015-12024.
doi: 10.1093/nar/gkx880 URL |
| [14] |
Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay[J]. Mol Microbiol, 2006, 60(3):723-737.
pmid: 16629673 |
| [15] |
Chen H, Previero A, Deutscher MP. A novel mechanism of ribonuclease regulation:GcvB and Hfq stabilize the mRNA that encodes RNase BN/Z during exponential phase[J]. J Biol Chem, 2019, 294(52):19997-20008.
doi: 10.1074/jbc.RA119.011367 pmid: 31744883 |
| [16] |
Bandyra KJ, Luisi BF. RNase E and the high-fidelity orchestration of RNA metabolism[J]. Microbiol Spectr, 2018, 6(2). DOI: 10.1128/microbiolspec.rwr-0008-2017.
doi: 10.1128/microbiolspec.rwr-0008-2017 |
| [17] |
Ali N, Gowrishankar J. Cross-subunit catalysis and a new phenomenon of recessive resurrection in Escherichia coli RNase E[J]. Nucleic Acids Res, 2020, 48(2):847-861.
doi: 10.1093/nar/gkz1152 URL |
| [18] |
Hadjeras L, Poljak L, Bouvier M, et al. Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RNase E and increased turnover of ribosome-free transcripts[J]. Mol Microbiol, 2019, 111(6):1715-1731.
doi: 10.1111/mmi.14248 pmid: 30903628 |
| [19] | Mardle CE, Goddard LR, Spelman BC, et al. Identification and analysis of novel small molecule inhibitors of RNase E:Implications for antibacterial targeting and regulation of RNase E[J]. Biochem Biophys Rep, 2020, 23:100773. |
| [20] |
Moore CJ, Go H, Shin E, et al. Substrate-dependent effects of quaternary structure on RNase E activity[J]. Genes Dev, 2021, 35(3/4):286-299.
doi: 10.1101/gad.335828.119 URL |
| [21] |
Hamouche L, Poljak L, Carpousis AJ. Ribosomal RNA degradation induced by the bacterial RNA polymerase inhibitor rifampicin[J]. bioRxiv, 2021. DOI: 10.1101/2021.04.24.441238.
doi: 10.1101/2021.04.24.441238 |
| [22] |
Mohanty BK, Agrawal A, Kushner SR. Generation of pre-tRNAs from polycistronic operons is the essential function of RNase P in Escherichia coli[J]. Nucleic Acids Res, 2020, 48(5):2564-2578.
doi: 10.1093/nar/gkz1188 pmid: 31993626 |
| [23] |
Kurata T, Nakanishi S, Hashimoto M, et al. Subunit composition of ribosome in the yqgF mutant Is deficient in pre-16S rRNA processing of Escherichia coli[J]. J Mol Microbiol Biotechnol, 2018, 28(4):179-182.
doi: 10.1159/000494494 URL |
| [24] |
Kouzminova EA, Kuzminov A. Ultraviolet-induced RNA:DNA hybrids interfere with chromosomal DNA synjournal[J]. Nucleic Acids Res, 2021, 49(7):3888-3906.
doi: 10.1093/nar/gkab147 pmid: 33693789 |
| [25] |
Prossliner T, Gerdes K, Sørensen MA, et al. Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation[J]. Nucleic Acids Res, 2021, 49(4):2226-2239.
doi: 10.1093/nar/gkab017 pmid: 33503254 |
| [26] |
Garcia-Rodriguez G, Charlier D, Wilmaerts D, et al. Alternative dimerization is required for activity and inhibition of the HEPN ribonuclease RnlA[J]. Nucleic Acids Res, 2021, 49(12):7164-7178.
doi: 10.1093/nar/gkab513 pmid: 34139012 |
| [27] | Dendooven T, Sinha D, Roeselová A, et al. A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation[J]. Mol Cell, 2021, 81(14):2901- 2913. e5. |
| [28] |
Dressaire C, Pobre V, Laguerre S, et al. PNPase is involved in the coordination of mRNA degradation and expression in stationary phase cells of Escherichia coli[J]. BMC Genomics, 2018, 19(1):848.
doi: 10.1186/s12864-018-5259-8 URL |
| [29] |
Park H, Yakhnin H, Connolly M, et al. CsrA participates in a PNPase autoregulatory mechanism by selectively repressing translation of pnp transcripts that have been previously processed by RNase III and PNPase[J]. J Bacteriol, 2015, 197(24):3751-3759.
doi: 10.1128/JB.00721-15 URL |
| [30] |
Fontaine F, Gasiorowski E, Gracia C, et al. The small RNA SraG participates in PNPase homeostasis[J]. RNA, 2016, 22(10):1560-1573.
doi: 10.1261/rna.055236.115 pmid: 27495318 |
| [31] |
Cairrão F, Chora A, Zilhão R, et al. RNase II levels change according to the growth conditions:characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II[J]. Mol Microbiol, 2001, 39(6):1550-1561.
pmid: 11260472 |
| [32] |
Lu F, Taghbalout A. Membrane association via an amino-terminal amphipathic helix is required for the cellular organization and function of RNase II[J]. J Biol Chem, 2013, 288(10):7241-7251.
doi: 10.1074/jbc.M112.408674 URL |
| [33] |
Song L, Wang G, Malhotra A, et al. Reversible acetylation on Lys501 regulates the activity of RNase II[J]. Nucleic Acids Res, 2016, 44(5):1979-1988.
doi: 10.1093/nar/gkw053 URL |
| [34] | dos Santos RF, Andrade JM, Pissarra J, et al. Hfq and RNase R mediate rRNA processing and degradation in a novel RNA quality control process[J]. mBio, 2020, 11(5):e02398-20. |
| [35] |
Liang W, Deutscher MP. Ribosomes regulate the stability and action of the exoribonuclease RNase R[J]. J Biol Chem, 2013, 288(48):34791-34798.
doi: 10.1074/jbc.M113.519553 URL |
| [36] |
Martínez VP, Dehò G, Simons RW, et al. Ribonuclease PH interacts with an acidic ribonuclease E site through a basic 80-amino acid domain[J]. FEMS Microbiol Lett, 2014, 355(1):51-60.
doi: 10.1111/fml.2014.355.issue-1 URL |
| [37] |
Sulthana S, Quesada E, Deutscher MP. RNase II regulates RNase PH and is essential for cell survival during starvation and stationary phase[J]. RNA, 2017, 23(9):1456-1464.
doi: 10.1261/rna.060558.116 pmid: 28625967 |
| [38] |
Zuo Y, Deutscher MP. The physiological role of RNase T can be explained by its unusual substrate specificity[J]. J Biol Chem, 2002, 277(33):29654-29661.
doi: 10.1074/jbc.M204252200 URL |
| [39] |
Jain C. RNase AM, a 5' to 3' exonuclease, matures the 5' end of all three ribosomal RNAs in E. coli[J]. Nucleic Acids Res, 2020, 48(10):5616-5623.
doi: 10.1093/nar/gkaa260 URL |
| [40] |
Mackie GA. RNase E:at the interface of bacterial RNA processing and decay[J]. Nat Rev Microbiol, 2013, 11(1):45-57.
doi: 10.1038/nrmicro2930 URL |
| [41] |
Mohanty BK, Petree JR, Kushner SR. Endonucleolytic cleavages by RNase E generate the mature 3' termini of the three proline tRNAs in Escherichia coli[J]. Nucleic Acids Res, 2016, 44(13):6350-6362.
doi: 10.1093/nar/gkw517 pmid: 27288443 |
| [42] |
Sulthana S, Basturea GN, Deutscher MP. Elucidation of pathways of ribosomal RNA degradation:an essential role for RNase E[J]. RNA, 2016, 22(8):1163-1171.
doi: 10.1261/rna.056275.116 pmid: 27298395 |
| [43] |
Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli[J]. Genes Dev, 2002, 16(9):1102-1115.
doi: 10.1101/gad.983502 URL |
| [44] |
Sousa S, Marchand I, Dreyfus M. Autoregulation allows Escherichia coli RNase E to adjust continuously its synjournal to that of its substrates[J]. Mol Microbiol, 2001, 42(3):867-878.
pmid: 11722748 |
| [45] |
Jain C, Deana A, Belasco JG. Consequences of RNase E scarcity in Escherichia coli[J]. Mol Microbiol, 2002, 43(4):1053-1064.
doi: 10.1046/j.1365-2958.2002.02808.x URL |
| [46] |
Mohanty BK, Kushner SR. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E[J]. Mol Microbiol, 2002, 45(5):1315-1324.
pmid: 12207699 |
| [47] |
Jaso-Vera ME, Domínguez-Malfavón L, Curiel-Quesada E, et al. Dynamics of the canonical RNA degradosome components during glucose stress[J]. Biochimie, 2021, 187:67-74.
doi: 10.1016/j.biochi.2021.05.006 pmid: 34022290 |
| [48] |
McQuail J, Carpousis AJ, Wigneshweraraj S. The association between Hfq and RNase E in long-term nitrogen starved Escherichia coli[J]. bioRxiv, 2021. DOI: 10.1101/2021.04, 19. 440462.
doi: 10.1101/2021.04 |
| [49] |
Singh D, Murashko ON, Lin-Chao SE. Posttranscriptional regulation of tnaA by protein-RNA interaction mediated by ribosomal protein L4 in Escherichia coli[J]. J Bacteriol, 2020, 202(10):e00799-19. DOI: 10.1128/jb.00799-19.
doi: 10.1128/jb.00799-19 |
| [50] |
Lee K, Zhan X, Gao J, et al. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli[J]. Cell, 2003, 114(5):623-634.
doi: 10.1016/j.cell.2003.08.003 URL |
| [51] | Lim B, Sim M, Lee H, et al. Regulation of Escherichia coli RNase III activity[J]. J Microbiol Seoul Korea, 2015, 53(8):487-494. |
| [52] |
Sirdeshmukh R, Schlessinger D. Ordered processing of Escherichia coli 23S rRNA in vitro[J]. Nucleic Acids Res, 1985, 13(14):5041-5054.
pmid: 2991850 |
| [53] |
King TC, Sirdeshmukh R, Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA[J]. PNAS, 1984, 81(1):185-188.
pmid: 6364133 |
| [54] |
Stead MB, Marshburn S, Mohanty BK, et al. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays[J]. Nucleic Acids Res, 2011, 39(8):3188-3203.
doi: 10.1093/nar/gkq1242 pmid: 21149258 |
| [55] |
Conrad C, Rauhut R. Ribonuclease III:new sense from nuisance[J]. Int J Biochem Cell Biol, 2002, 34(2):116-129.
doi: 10.1016/S1357-2725(01)00112-1 URL |
| [56] |
Deutscher MP. Regulation of bacterial ribonucleases[J]. Annu Rev Microbiol, 2021, 75(1). DOI: 10.1146/annurev-micro-020121-011201.
doi: 10.1146/annurev-micro-020121-011201 |
| [57] |
Lee J, Lee M, Lee K. Trans-acting regulators of ribonuclease activity[J]. J Microbiol, 2021, 59(4):341-359.
doi: 10.1007/s12275-021-0650-6 URL |
| [58] |
Paudyal S, Alfonso-Prieto M, Carnevale V, et al. Combined computational and experimental analysis of a complex of ribonuclease III and the regulatory macrodomain protein, YmdB[J]. Proteins, 2015, 83(3):459-472.
doi: 10.1002/prot.v83.3 URL |
| [59] |
Kavalchuk K, Madhusudan S, Schnetz K. RNase III initiates rapid degradation of proU mRNA upon hypo-osmotic stress in Escherichia coli[J]. RNA Biol, 2012, 9(1):98-109.
doi: 10.4161/rna.9.1.18228 URL |
| [60] |
Song W, Kim YH, Sim SH, et al. Antibiotic stress-induced modulation of the endoribonucleolytic activity of RNase III and RNase G confers resistance to aminoglycoside antibiotics in Escherichia coli[J]. Nucleic Acids Res, 2014, 42(7):4669-4681.
doi: 10.1093/nar/gku093 URL |
| [61] |
Li de la Sierra-Gallay I, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z[J]. Nature, 2005, 433(7026):657-661.
doi: 10.1038/nature03284 URL |
| [62] |
Chen H, Dutta T, Deutscher MP. Growth phase-dependent variation of RNase BN/Z affects small RNAs[J]. J Biol Chem, 2016, 291(51):26435-26442.
pmid: 27875308 |
| [63] |
Gupta AK, Siddiqui N, Dutta T. A novel mechanism of RyeA/SraC induction under acid stress[J]. Biochem Biophys Res Commun, 2020, 525(2):298-302.
doi: 10.1016/j.bbrc.2020.02.085 URL |
| [64] |
Grünberg S, Coxam B, Chen TH, et al. E. coli RNase I exhibits a strong Ca2+-dependent inherent double-stranded RNase activity[J]. Nucleic Acids Res, 2021, 49(9):5265-5277.
doi: 10.1093/nar/gkab284 URL |
| [65] |
Fontaine BM, Martin KS, Garcia-Rodriguez JM, et al. RNase I regulates Escherichia coli 2', 3'-cyclic nucleotide monophosphate levels and biofilm formation[J]. Biochem J, 2018, 475(8):1491-1506.
doi: 10.1042/BCJ20170906 URL |
| [66] |
Hossain ST, Malhotra A, Deutscher MP. How RNase R degrades structured rna:role of the helicase activity and the S1 domain[J]. J Biol Chem, 2016, 291(15):7877-7887.
doi: 10.1074/jbc.M116.717991 URL |
| [67] |
Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II[J]. J Biol Chem, 2002, 277(24):21624-21629.
doi: 10.1074/jbc.M202942200 URL |
| [68] |
Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions[J]. J Biol Chem, 2005, 280(41):34393-34396.
doi: 10.1074/jbc.C500333200 URL |
| [69] |
Liang WX, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein:growth stage-dependent modification of RNase R[J]. Mol Cell, 2011, 44(1):160-166.
doi: 10.1016/j.molcel.2011.06.037 URL |
| [70] |
Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3' right-arrow 5' exonuclease and a poly(A)polymerase in Escherichiacoli[J]. PNAS, 2000, 97(22):11966-11971.
pmid: 11035800 |
| [71] |
Briani F, Carzaniga T, Dehò G. Regulation and functions of bacterial PNPase[J]. Wiley Interdiscip Rev RNA, 2016, 7(2):241-258.
doi: 10.1002/wrna.2016.7.issue-2 URL |
| [72] |
Cameron TA, de Lay NR. The phosphorolytic exoribonucleases polynucleotide phosphorylase and RNase PH stabilize sRNAs and facilitate regulation of their mRNA targets[J]. J Bacteriol, 2016, 198(24):3309-3317.
pmid: 27698082 |
| [73] |
Saramago M, Bárria C, dos Santos RF, et al. The role of RNases in the regulation of small RNAs[J]. Curr Opin Microbiol, 2014, 18:105-115.
doi: 10.1016/j.mib.2014.02.009 URL |
| [74] |
Pobre V, Arraiano CM. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli[J]. BMC Genomics, 2015, 16:72.
doi: 10.1186/s12864-015-1237-6 URL |
| [75] |
Wu J, Jiang Z, Liu M, et al. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress[J]. Biochemistry, 2009, 48(9):2012-2020.
doi: 10.1021/bi801752p URL |
| [76] |
Hör J, Matera G, Vogel J, et al. Trans-acting small RNAs and their effects on gene expression in Escherichia coli and Salmonella enterica[J]. Ecosal Plus, 2020,9:ecosalplus. ESP-30. DOI: 10.1128/ecosalplus.esp-0030-2019.
doi: 10.1128/ecosalplus.esp-0030-2019 |
| [77] |
Nurmohamed S, Vincent HA, Titman CM, et al. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli[J]. J Biol Chem, 2011, 286(16):14315-14323.
doi: 10.1074/jbc.M110.200741 pmid: 21324911 |
| [78] | Arraiano CM, Mauxion F, Viegas SC, et al. Intracellular ribonucleases involved in transcript processing and decay:Precision tools for RNA[J]. Biochim et Biophys Acta BBA Gene Regul Mech, 2013, 1829(6/7):491-513. |
| [79] |
Mohanty BK, Maples VF, Kushner SR. Polyadenylation helps regulate functional tRNA levels in Escherichia coli[J]. Nucleic Acids Res, 2012, 40(10):4589-4603.
doi: 10.1093/nar/gks006 pmid: 22287637 |
| [80] |
Garza-Sánchez F, Shoji S, Fredrick K, et al. RNase II is important for A-site mRNA cleavage during ribosome pausing[J]. Mol Microbiol, 2009, 73(5):882-897.
doi: 10.1111/j.1365-2958.2009.06813.x pmid: 19627501 |
| [81] |
Pobre V, Barahona S, Dobrzanski T, et al. Defining the impact of exoribonucleases in the shift between exponential and stationary phases[J]. Sci Rep, 2019, 9(1):16271.
doi: 10.1038/s41598-019-52453-6 URL |
| [82] |
Gutgsell NS, Jain C. Role of precursor sequences in the ordered maturation of E. coli 23S ribosomal RNA[J]. RNA, 2012, 18(2):345-353.
doi: 10.1261/rna.027854.111 pmid: 22190745 |
| [83] |
Li Z, Pandit S, Deutscher MP. 3' exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli[J]. PNAS, 1998, 95(6):2856-2861.
pmid: 9501180 |
| [1] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [2] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [3] | TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates [J]. Biotechnology Bulletin, 2023, 39(11): 205-216. |
| [4] | LI Hai-li, LANG Li-min, ZHANG Qing-xian, YOU Yi, ZHU Wen-hao, WANG Zhi-fang, ZHANG Li-xian, WANG Ke-ling. Identification and Drug Resistance of Escherichia coli Simultaneously Producing Carbapenemase NDM-1 and NDM-5 [J]. Biotechnology Bulletin, 2022, 38(9): 106-115. |
| [5] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [6] | ZHAO Yan-kun, LIU Hui-min, MENG Lu, WANG Cheng, WANG Jia-qi, ZHENG Nan. Research Progress in Heteroresistance of Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(9): 59-71. |
| [7] | WEI Xin-xin, LAN Hai-yan. Advances in the Regulation of Plant MYB Transcription Factors in Secondary Metabolism and Stress Response [J]. Biotechnology Bulletin, 2022, 38(8): 12-23. |
| [8] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [9] | WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(12): 252-264. |
| [10] | ZOU Kun, LU Li-li, Collins Asiamah Amponsah, XUE Yuan, ZHANG Shao-wei, SU Ying, ZHAO Zhi-hui. Research Progress on Mechanism of Poultry Follicular Atresia [J]. Biotechnology Bulletin, 2020, 36(4): 185-191. |
| [11] | WEI Ming-ming, ZENG Xia, AN Ze-wei, HU Yan-shi, HUANG Xiao, LI Wei-guo. Advances in the Maintenance and Termination of Floral Meristem Regulated by C-type Floral Organ Gene AGAMOUS(AG) [J]. Biotechnology Bulletin, 2020, 36(1): 135-143. |
| [12] | LI Xiao-yuan, XIE Li-nan. Research Progress in Na+ Regulation Mechanism of Plants Under Salt Stress [J]. Biotechnology Bulletin, 2019, 35(7): 148-155. |
| [13] | KUANG Yong-jie, LIU Lang, YAN Fang, REN Bin, YAN Da-qi, ZHANG Da-wei, LIN Hong-hui, ZHOU Huan-bin. Functions of Phytohormones During the Interactions Between Rice, Pathogens [J]. Biotechnology Bulletin, 2018, 34(2): 74-86. |
| [14] | FENG Zhi-mei, ZHAO Ya-tong, LIU Ye-xue, LU Fu-ping, LI Yu. Recombinant Expression of NADPH-Dependent Mannitol Dehydrogenase and Transformation Conditions of Mannitol [J]. Biotechnology Bulletin, 2017, 33(8): 186-191. |
| [15] | LIU Xiao-wei, YANG Xiu-yan, LIU Zheng-xiang, WU Hai-wen, ZHANG Hua-xin, ZHU Jian-feng. Role of MicroRNA in Plant Resistance to Salt Stress [J]. Biotechnology Bulletin, 2017, 33(12): 12-21. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||