








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (2): 126-138.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0567
Previous Articles Next Articles
MU De-tian1( ), WAN Ling-yun2, ZHANG Yao1, WEI Shu-gen2, LU Ying1, FU Jin-e2, TIAN Yi1, PAN Li-mei2(
), WAN Ling-yun2, ZHANG Yao1, WEI Shu-gen2, LU Ying1, FU Jin-e2, TIAN Yi1, PAN Li-mei2( ), TANG Qi1(
), TANG Qi1( )
)
Received:2022-05-09
Online:2023-02-26
Published:2023-03-07
MU De-tian, WAN Ling-yun, ZHANG Yao, WEI Shu-gen, LU Ying, FU Jin-e, TIAN Yi, PAN Li-mei, TANG Qi. House-keeping Genes Screening and Expression Patterns Analysis of Genes Involved in Alkaloid Biosynthesis in Uncaria rhynchophylla[J]. Biotechnology Bulletin, 2023, 39(2): 126-138.
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| G8H | Geraniol 8-hydroxylase | F:CGCCCATAATCATTCCACTA R:GCCAATGTCACTGCCTCAC |
| 8-HGO | 8-hydroxygeraniol oxidoreductase | F:GCGTCTGGTGTTCTATCTCCGTT R:GGTTGCGTTGCTGCCTACAT |
| IS | Iridoid synthase | F:ACCCTTCAGAAACTCAACAAAAGC R:CCTCGCAACATTCTGGGCTA |
| CYP76A26 | Nepetalactol monooxygenase | F:ACCTTATGCTGTCGAGGGATT R:TAACACTGCCAGCTTTCCTGT |
| 7-DLGT / UGT85A24 | 7-deoxyloganetic acid glucosyltransferase | F:CATGGAAGAAGACAGCAGGAG R:CATTGTCGAATTCCAACCACT |
| 7-DLH / CYP2A224 | 7-deoxyloganate 7-hydroxylase | F:AGGAACAACAGGATGAGAGCA R:AGGTAGTGTCCTGTCCAGCAA |
| LAMT | Loganate methyltransferase | F:TGTCAAAGAAATGCCTGAAGAAG R:ATGCGGAAAGGCTTAGATGG |
| SLS / CYP72A1 | Secologanin synthase | F:TCGGACCATTCAAACCTACGG R:GCTTGGACCATCTGTCACCCTC |
| AS | Anthranilate synthase | F:TCAAGGACGAAGGGTGGAACAG R:ACCAACCCAACCACCACAAAAT |
| AnPRT | Anthranilate phosphoribosyltransferase | F:GTTGGGACTGGTGGTGATGG R:CTTGACCTATTTCCTTGCTTTGC |
| IGPS | Indole-3-glycerol phosphate synthase | F:GTTGGGGAGTCTGGGCTTTT R:GAGTCCGGTGATTGCCTTAGTT |
| TSA | Tryptophan synthase alpha chain | F:TGTGAAACAAGTTGCTGGATGG R:AGGAGATTTTGCCTCGCCTA |
| TSB | Tryptophan synthase beta chain | F:GCTGAGGTTAGGCCAGTTCATT R:TCCACATTAGTCACCCAGTCCC |
| TDC | L-tryptophan decarboxylase | F:GGCAGGTATTTTCCCACGCA R:GTTCCCACGGTAGCACAGA |
| STR | Strictosidine synthase | F:GCCGATGGTCGGATTCTCAA R:GGCCCAAATAGGCATCAGCA |
Table 1 Primer sequences for key genes in U. rhynchophylla alkaloids upstream synthesis pathway for RT-qPCR
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| G8H | Geraniol 8-hydroxylase | F:CGCCCATAATCATTCCACTA R:GCCAATGTCACTGCCTCAC |
| 8-HGO | 8-hydroxygeraniol oxidoreductase | F:GCGTCTGGTGTTCTATCTCCGTT R:GGTTGCGTTGCTGCCTACAT |
| IS | Iridoid synthase | F:ACCCTTCAGAAACTCAACAAAAGC R:CCTCGCAACATTCTGGGCTA |
| CYP76A26 | Nepetalactol monooxygenase | F:ACCTTATGCTGTCGAGGGATT R:TAACACTGCCAGCTTTCCTGT |
| 7-DLGT / UGT85A24 | 7-deoxyloganetic acid glucosyltransferase | F:CATGGAAGAAGACAGCAGGAG R:CATTGTCGAATTCCAACCACT |
| 7-DLH / CYP2A224 | 7-deoxyloganate 7-hydroxylase | F:AGGAACAACAGGATGAGAGCA R:AGGTAGTGTCCTGTCCAGCAA |
| LAMT | Loganate methyltransferase | F:TGTCAAAGAAATGCCTGAAGAAG R:ATGCGGAAAGGCTTAGATGG |
| SLS / CYP72A1 | Secologanin synthase | F:TCGGACCATTCAAACCTACGG R:GCTTGGACCATCTGTCACCCTC |
| AS | Anthranilate synthase | F:TCAAGGACGAAGGGTGGAACAG R:ACCAACCCAACCACCACAAAAT |
| AnPRT | Anthranilate phosphoribosyltransferase | F:GTTGGGACTGGTGGTGATGG R:CTTGACCTATTTCCTTGCTTTGC |
| IGPS | Indole-3-glycerol phosphate synthase | F:GTTGGGGAGTCTGGGCTTTT R:GAGTCCGGTGATTGCCTTAGTT |
| TSA | Tryptophan synthase alpha chain | F:TGTGAAACAAGTTGCTGGATGG R:AGGAGATTTTGCCTCGCCTA |
| TSB | Tryptophan synthase beta chain | F:GCTGAGGTTAGGCCAGTTCATT R:TCCACATTAGTCACCCAGTCCC |
| TDC | L-tryptophan decarboxylase | F:GGCAGGTATTTTCCCACGCA R:GTTCCCACGGTAGCACAGA |
| STR | Strictosidine synthase | F:GCCGATGGTCGGATTCTCAA R:GGCCCAAATAGGCATCAGCA |
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') | 斜率k | 扩增效率E/% | 相关系数R2 |
|---|---|---|---|---|---|
| 18S | 18S Ribosomal RNA | F:CTTCGGGATCGGAGTAATGA R:GCGGAGTCCTAGAAGCAACA | -3.14 | 0.97 | 0.999 96 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:CAGGAACCCAGAGGAGATT R:GCATCCTTACTTGGAGCAG | -3.41 | 0.96 | 0.999 78 |
| Actin6 | Actin 6 | F:ACCGAGCGTGGTTATTCTT R:TTCCTGCTGCTTCCATTCC | -3.54 | 0.92 | 0.991 79 |
| EF1-β | Elongation factor 1β | F:AAGGCATCCACCAAGAAGA R:AAGGCAACAATGTCACAGC | -3.49 | 0.93 | 0.998 07 |
| TUB | β-Tubulin | F:GGAAGTAATCTGCGACGAG R:TGAGAACAGCACGAGGGAC | -3.12 | 1.09 | 0.998 71 |
| CYP | Cyclophilin | F:CGAGAAAGGCGTGGGAAAG R:TGAGACCCGTTGGTGTTGG | -3.2 | 1.05 | 0.998 35 |
| EF1-α | Elongation factor 1α | F:GAGCGTGAGCGTGGTATTACTAT R:CCAGTGGTGGAGTCAATGATAA | -3.12 | 1.09 | 0.994 50 |
| PAL | Phenylalanine ammonialyase | F:ATCGCTGAATCCTCCAATA R:CCACCCTACTCCACAATACTT | -3.12 | 1.09 | 0.998 68 |
| RNA L13 | Ribosomal protein L13 | F:CCAGGAGAAGGAATGCGAGG R:GACCGGTTTTTACGGCGATG | -3.67 | 0.87 | 0.991 99 |
| SAM | S-adenosylmethionine decarboxylase | F:CACAATCTGGCATACGAAA R:AACTCACTTGGCTGGAAAC | -3.7 | 0.92 | 0.995 17 |
| cdc73 | Cell division control protein 73 | F:TGGTGGCTGTTTTCGTGTT R:TGATGCCGCTTATTCTTGC | -3.44 | 0.95 | 0.990 33 |
| TUA | α-Tubulin | F:TCCCTTCTTGAGCACACTGAT R:CCATCAAACCTCAAAGACGCA | -3.28 | 1.02 | 0.993 96 |
Table 2 Primer sequence and amplification parameters for 12 candidate reference genes of U. rhynchophylla in RT-qPCR
| 基因Gene | 基因全称Gene name | 引物序列Primer sequence(5'-3') | 斜率k | 扩增效率E/% | 相关系数R2 |
|---|---|---|---|---|---|
| 18S | 18S Ribosomal RNA | F:CTTCGGGATCGGAGTAATGA R:GCGGAGTCCTAGAAGCAACA | -3.14 | 0.97 | 0.999 96 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F:CAGGAACCCAGAGGAGATT R:GCATCCTTACTTGGAGCAG | -3.41 | 0.96 | 0.999 78 |
| Actin6 | Actin 6 | F:ACCGAGCGTGGTTATTCTT R:TTCCTGCTGCTTCCATTCC | -3.54 | 0.92 | 0.991 79 |
| EF1-β | Elongation factor 1β | F:AAGGCATCCACCAAGAAGA R:AAGGCAACAATGTCACAGC | -3.49 | 0.93 | 0.998 07 |
| TUB | β-Tubulin | F:GGAAGTAATCTGCGACGAG R:TGAGAACAGCACGAGGGAC | -3.12 | 1.09 | 0.998 71 |
| CYP | Cyclophilin | F:CGAGAAAGGCGTGGGAAAG R:TGAGACCCGTTGGTGTTGG | -3.2 | 1.05 | 0.998 35 |
| EF1-α | Elongation factor 1α | F:GAGCGTGAGCGTGGTATTACTAT R:CCAGTGGTGGAGTCAATGATAA | -3.12 | 1.09 | 0.994 50 |
| PAL | Phenylalanine ammonialyase | F:ATCGCTGAATCCTCCAATA R:CCACCCTACTCCACAATACTT | -3.12 | 1.09 | 0.998 68 |
| RNA L13 | Ribosomal protein L13 | F:CCAGGAGAAGGAATGCGAGG R:GACCGGTTTTTACGGCGATG | -3.67 | 0.87 | 0.991 99 |
| SAM | S-adenosylmethionine decarboxylase | F:CACAATCTGGCATACGAAA R:AACTCACTTGGCTGGAAAC | -3.7 | 0.92 | 0.995 17 |
| cdc73 | Cell division control protein 73 | F:TGGTGGCTGTTTTCGTGTT R:TGATGCCGCTTATTCTTGC | -3.44 | 0.95 | 0.990 33 |
| TUA | α-Tubulin | F:TCCCTTCTTGAGCACACTGAT R:CCATCAAACCTCAAAGACGCA | -3.28 | 1.02 | 0.993 96 |
| 基因 Gene | 几何平均数 GM | 算术平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 Standard deviation | 变异系数 Coefficient of variation | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|---|
| 18S | 15.55 | 15.55 | 15.09 | 16.15 | 0.32 | 2.05 | 1 |
| SAM | 22.14 | 22.15 | 21.10 | 23.05 | 0.54 | 2.44 | 2 |
| CYP | 19.39 | 19.41 | 18.05 | 20.77 | 0.69 | 3.56 | 3 |
| EF-1β | 21.58 | 21.59 | 20.43 | 22.68 | 0.75 | 3.48 | 4 |
| cdc73 | 24.46 | 24.48 | 23.35 | 26.36 | 0.77 | 3.15 | 5 |
| TUB | 24.30 | 24.31 | 23.42 | 25.68 | 0.78 | 3.22 | 6 |
| EF-1α | 21.05 | 21.07 | 19.34 | 22.51 | 0.87 | 4.12 | 7 |
| PAL | 22.94 | 22.96 | 21.20 | 23.91 | 0.96 | 4.16 | 8 |
| Actin6 | 23.46 | 23.51 | 21.11 | 24.90 | 1.17 | 4.91 | 9 |
| RNA L13 | 22.50 | 22.54 | 21.03 | 25.07 | 1.18 | 5.21 | 10 |
| GAPDH | 26.82 | 26.87 | 24.55 | 28.70 | 1.53 | 5.69 | 11 |
| TUA | 22.36 | 22.53 | 19.15 | 26.03 | 2.78 | 12.34 | 12 |
Table 3 Expression stabilities of 12 candidate reference genes analyzed by BestKeeper
| 基因 Gene | 几何平均数 GM | 算术平均数 AM | 最小值 Min | 最大值 Max | 标准偏差 Standard deviation | 变异系数 Coefficient of variation | 稳定性排序 Stability rank |
|---|---|---|---|---|---|---|---|
| 18S | 15.55 | 15.55 | 15.09 | 16.15 | 0.32 | 2.05 | 1 |
| SAM | 22.14 | 22.15 | 21.10 | 23.05 | 0.54 | 2.44 | 2 |
| CYP | 19.39 | 19.41 | 18.05 | 20.77 | 0.69 | 3.56 | 3 |
| EF-1β | 21.58 | 21.59 | 20.43 | 22.68 | 0.75 | 3.48 | 4 |
| cdc73 | 24.46 | 24.48 | 23.35 | 26.36 | 0.77 | 3.15 | 5 |
| TUB | 24.30 | 24.31 | 23.42 | 25.68 | 0.78 | 3.22 | 6 |
| EF-1α | 21.05 | 21.07 | 19.34 | 22.51 | 0.87 | 4.12 | 7 |
| PAL | 22.94 | 22.96 | 21.20 | 23.91 | 0.96 | 4.16 | 8 |
| Actin6 | 23.46 | 23.51 | 21.11 | 24.90 | 1.17 | 4.91 | 9 |
| RNA L13 | 22.50 | 22.54 | 21.03 | 25.07 | 1.18 | 5.21 | 10 |
| GAPDH | 26.82 | 26.87 | 24.55 | 28.70 | 1.53 | 5.69 | 11 |
| TUA | 22.36 | 22.53 | 19.15 | 26.03 | 2.78 | 12.34 | 12 |
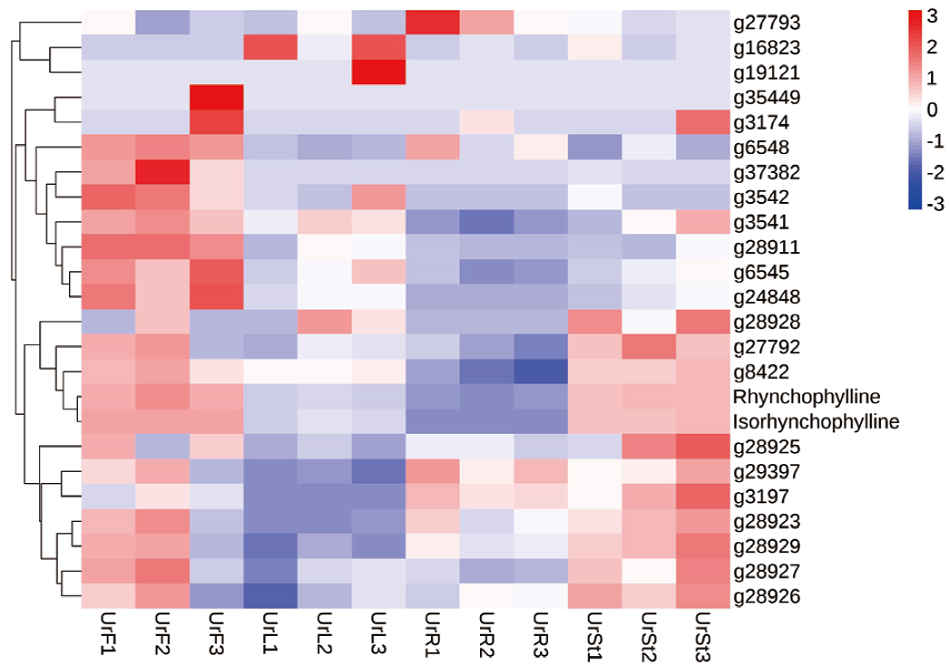
Fig. 9 Screening heatmap of strictosidine synthase gene Rhynchophylline is the content of rhynchophylline, and isorhynchophylline is the content of isorhynchophylline
| [1] | 国家药典委员会. 中国药典[M]. 北京: 中国医药科技出版社, 2020, 268. |
| Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China.[M]. Beijing: China Medical Science Press, 2020: 268. | |
| [2] | 高晓宇, 丁茹, 王道平, 等. 钩藤化学成分及药理作用研究进展[J]. 天津医科大学学报, 2017, 23(4): 380-382. |
| Gao XY, Ding R, Wang DP, et al. Advances in chemical constituents and pharmacological action of Uncaria rhynchophylla.[J]. J Tianjin Med Univ, 2017, 23(4): 380-382. | |
| [3] | 顾雄华, 刘芊, 贺小芳. 天麻钩藤饮合小柴胡汤治疗围绝经期高血压合并焦虑状态临床观察[J]. 北京中医药, 2012, 31(4): 305-308. |
| Gu XH, Liu Q, He XF. Clinical observation of tianma gouteng decoction combined with xiaochaihu decoction in the treatment of perimenopausal hypertension combined with anxiety[J]. Beijing J Tradit Chin Med, 2012, 31(4): 305-308. | |
| [4] | 周吉银, 周世文, 贺燕. 异钩藤碱药理作用的最新研究进展[J]. 中成药, 2013, 35(3): 596-599. |
| Zhou JY, Zhou SW, He Y. The latest research progress of pharmacological action of Isorhynchophylline[J]. Chin Tradit Pat Med, 2013, 35(3): 596-599. | |
| [5] |
Li HL, Wang XB, Liu Y, et al. Hepatoprotection and hepatotoxicity of Heshouwu, a Chinese medicinal herb: context of the paradoxical effect[J]. Food Chem Toxicol, 2017, 108(Pt B): 407-418.
doi: S0278-6915(16)30264-2 pmid: 27484243 |
| [6] | 柳威, 邓林华, 赵英强. 钩藤提取物及钩藤碱的药理研究进展[J]. 中药新药与临床药理, 2021, 32(6): 899-904. |
| Liu W, Deng LH, Zhao YQ. Research progress on pharmacological effects of Uncaria extract and rhynchophylline[J]. Tradit Chin Drug Res Clin Pharmacol, 2021, 32(6): 899-904. | |
| [7] |
Pan HQ, Yang WZ, Zhang YB, et al. An integrated strategy for the systematic characterization and discovery of new indole alkaloids from Uncaria rhynchophylla by UHPLC/DAD/LTQ-Orbitrap-MS[J]. Anal Bioanal Chem, 2015, 407(20): 6057-6070.
doi: 10.1007/s00216-015-8777-0 URL |
| [8] |
Wei SG, Luo ZL, Cui SR, et al. Molecular identification and targeted quantitative analysis of medicinal materials from Uncaria species by DNA barcoding and LC-MS/MS[J]. Molecules, 2019, 24(1): 175.
doi: 10.3390/molecules24010175 URL |
| [9] |
Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays[J]. J Mol Endocrinol, 2000, 25(2): 169-193.
doi: 10.1677/jme.0.0250169 pmid: 11013345 |
| [10] |
Mahoney DJ, Carey K, Fu MH, et al. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise[J]. Physiol Genomics, 2004, 18(2): 226-231.
pmid: 15161965 |
| [11] |
Maroufi A, van Bockstaele E, de Loose M. Validation of reference genes for gene expression analysis in chicory(Cichorium intybus)using quantitative real-time PCR[J]. BMC Mol Biol, 2010, 11: 15.
doi: 10.1186/1471-2199-11-15 pmid: 20156357 |
| [12] | 易小哲, 邬兰, 向丽, 等. 艾Artemisia argyi实时荧光定量PCR内参基因筛选[J]. 中国中药杂志, 2022, 47(3): 659-667. |
| Yi XZ, Wu L, Xiang L, et al. Screening of reference genes for quantitative real-time PCR in Artemisia argyi[J]. China J Chin Mater Med, 2022, 47(3): 659-667. | |
| [13] | 涂冬萍, 莫长明, 马小军, 等. 罗汉果实时荧光定量PCR内参基因的选择[J]. 中国中药杂志, 2015, 40(2): 204-209. |
| Tu DP, Mo CM, Ma XJ, et al. Selection of reference genes of Siraitia grosvenorii by real-time PCR[J]. China J Chin Mater Med, 2015, 40(2): 204-209. | |
| [14] | 杨婷, 薛珍珍, 李娜, 等. 铁十字秋海棠斑叶发育过程内参基因筛选及验证[J]. 园艺学报, 2021, 48(11): 2251-2261. |
| Yang T, Xue ZZ, Li N, et al. Reference genes selection and validation in Begonia masoniana leaves of different developmental stages[J]. Acta Hortic Sin, 2021, 48(11): 2251-2261. | |
| [15] | 郭茜茜. 钩藤碱和异钩藤碱生物合成路径的研究[D]. 哈尔滨: 东北农业大学, 2014. |
| Guo QQ. Research on rhynchophylline and isorhynchophylline biosynthetic pathway[D]. Harbin: Northeast Agricultural University, 2014. | |
| [16] |
Contin A, van der Heijden R, Lefeber AW, et al. The iridoid glucoside secologanin is derived from the novel triose phosphate/pyruvate pathway in a Catharanthus roseus cell culture[J]. FEBS Lett, 1998, 434(3): 413-416.
doi: 10.1016/s0014-5793(98)01022-9 pmid: 9742965 |
| [17] |
Yamazaki Y, Kitajima M, Arita M, et al. Biosynthesis of camptothecin. in silico and in vivo tracer study from[1-13C]glucose[J]. Plant Physiol, 2004, 134(1): 161-170.
pmid: 14657405 |
| [18] |
Collu G, Unver N, Peltenburg-Looman AM, et al. Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis[J]. FEBS Lett, 2001, 508(2): 215-220.
doi: 10.1016/s0014-5793(01)03045-9 pmid: 11718718 |
| [19] |
Collu G, Alonso Garcia A, van der Heijden R, et al. Activity of the cytochrome P450 enzyme geraniol 10-hydroxylase and alkaloid production in plant cell cultures[J]. Plant Sci, 2002, 162(1): 165-172.
doi: 10.1016/S0168-9452(01)00554-4 URL |
| [20] |
Yamamoto H, Katano N, Ooi A, et al. Secologanin synthase which catalyzes the oxidative cleavage of loganin into secologanin is a cytochrome P450[J]. Phytochemistry, 2000, 53(1): 7-12.
pmid: 10656401 |
| [21] |
Irmler S, Schroder G, St-Pierre B, et al. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase[J]. Plant J, 2000, 24(6): 797-804.
doi: 10.1046/j.1365-313x.2000.00922.x pmid: 11135113 |
| [22] |
Stöckigt J, Barleben L, Panjikar S, et al. 3D-Structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis[J]. Plant Physiol Biochem, 2008, 46(3): 340-355.
doi: 10.1016/j.plaphy.2007.12.011 URL |
| [23] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7): RESEARCH0034. |
| [24] |
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15): 5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 pmid: 15289330 |
| [25] |
Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations[J]. Biotechnol Lett, 2004, 26(6): 509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [26] |
Xie F, Xiao P, Chen D, et al. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs[J]. Plant Mol Biol, 2012, 80(1): 75-84.
doi: 10.1007/s11103-012-9885-2 URL |
| [27] | 姚李祥, 潘春柳, 余丽莹, 等. 草果种子休眠解除过程中RT-qPCR内参基因筛选[J]. 中国中药杂志, 2021, 46(15): 3832-3837. |
| Yao LX, Pan CL, Yu LY, et al. Selection of RT-qPCR reference genes for Amomum tsaoko seeds during dormancy release[J]. China J Chin Mater Med, 2021, 46(15): 3832-3837. | |
| [28] | 吴建阳, 何冰, 杜玉洁, 等. 利用geNorm、NormFinder和BestKeeper软件进行内参基因稳定性分析的方法[J]. 现代农业科技, 2017(5): 278-281. |
| Wu JY, He B, Du YJ, et al. Analysis method of systematically evaluating stability of reference genes using geNorm, NormFinder and BestKeeper[J]. Mod Agric Sci Technol, 2017(5): 278-281. | |
| [29] |
Tang Q, Ma XJ, Mo CM, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis[J]. BMC Genomics, 2011, 12: 343.
doi: 10.1186/1471-2164-12-343 pmid: 21729270 |
| [30] |
Dai LH, Liu C, Zhu YM, et al. Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii[J]. Plant Cell Physiol, 2015, 56(6): 1172-1182.
doi: 10.1093/pcp/pcv043 URL |
| [31] |
Zhang JS, Dai LH, Yang JG, et al. Oxidation of cucurbitadienol catalyzed by CYP87D18 in the biosynthesis of mogrosides from Siraitia grosvenorii[J]. Plant Cell Physiol, 2016, 57(5): 1000-1007.
doi: 10.1093/pcp/pcw038 URL |
| [32] | 陈凌艳, 谢德金, 荣俊冬, 等. 花叶唐竹4种叶色表型RT-qPCR内参基因筛选[J]. 分子植物育种, 2019, 17(14): 4592-4599. |
| Chen LY, Xie DJ, Rong JD, et al. Screening of RT-qPCR internal reference genes for four leaf color phenotypes of Sinobambusa tootsik f. luteoloalbostriata[J]. Mol Plant Breed, 2019, 17(14): 4592-4599. | |
| [33] | 代红军, 秦晨亮, 徐伟荣. 赤霞珠葡萄发育后期RT-PCR内参基因的筛选和验证[J]. 江苏农业学报, 2016, 32(3): 668-673. |
| Dai HJ, Qin CL, Xu WR. Screening and validation of reference genes for real-time fluorescence quantitative PCR during grape berry development of Cabernet Sauvignon[J]. Jiangsu J Agric Sci, 2016, 32(3): 668-673. | |
| [34] | 杨坤, 黄超, 卢山, 等. 铜胁迫下紫鸭跖草根组织实时定量PCR内参基因的选择[J]. 植物生理学报, 2021, 57(1): 195-204. |
| Yang K, Huang C, Lu S, et al. Reference gene selection for quantitative real-time PCR in purple setcreasea(Setcreasea purpurea)root tissue under copper stress[J]. Plant Physiol J, 2021, 57(1): 195-204. | |
| [35] |
Geu-Flores F, Sherden NH, Courdavault V, et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis[J]. Nature, 2012, 492(7427): 138-142.
doi: 10.1038/nature11692 URL |
| [36] | 吴昕怡, 刘小莉. 环烯醚萜类成分生物合成途径及关键酶基因研究进展[J]. 中国民族民间医药, 2017, 26(8): 44-48. |
| Wu XY, Liu XL. Progress of biosynthetic pathway and the key enzyme genes of iridoids[J]. Chin J Ethnomedicine Ethnopharmacy, 2017, 26(8): 44-48. | |
| [37] |
Noé W, Mollenschott C, Berlin J. Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: purification, molecular and kinetic data of the homogenous protein[J]. Plant Mol Biol, 1984, 3(5): 281-288.
doi: 10.1007/BF00017782 pmid: 24310513 |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [3] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [4] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [7] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [8] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [9] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [10] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [11] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [12] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [13] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [14] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [15] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||