








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 255-267.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0086
Previous Articles Next Articles
MIAO Yong-mei1( ), MIAO Cui-ping2, YU Qing-cai1
), MIAO Cui-ping2, YU Qing-cai1
Received:2023-02-07
Online:2023-09-26
Published:2023-10-24
MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum[J]. Biotechnology Bulletin, 2023, 39(9): 255-267.
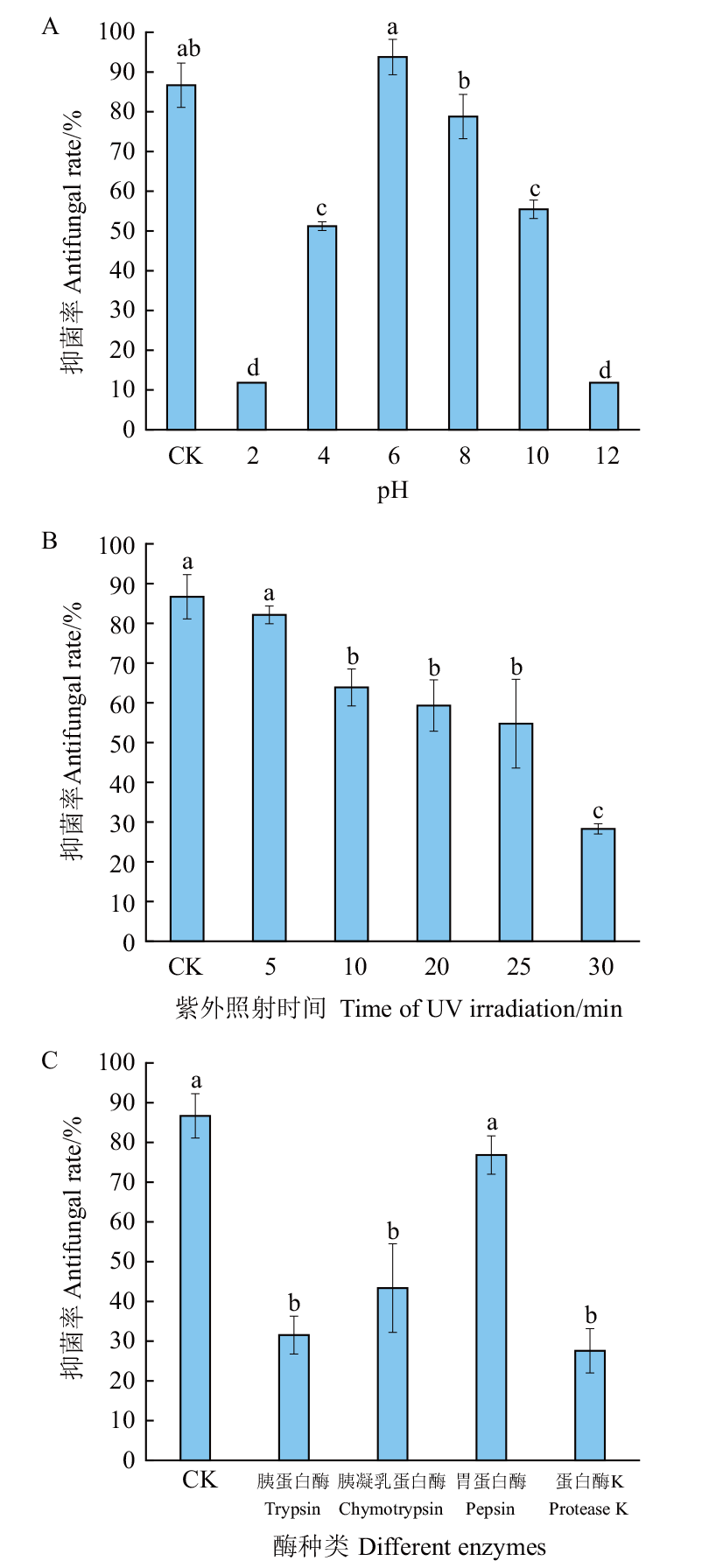
Fig. 2 Inhibitory effects of fermentation broths after different treatments Different lowercase letters indicate significant difference at 0.05 level. The same below
| 指标 Index | 抑菌率 Antifungal rate/% | SOD活力 SOD activity/ (U·g-1) | POD活力 POD activity/ (U·g-1) | CAT活力 CAT activity/ (U·g-1) | 可溶性蛋白含量 Soluble protein content/ (mg·g-1) | 可溶性糖含量 Soluble sugar content/ (mg·g-1) |
|---|---|---|---|---|---|---|
| 抑菌率Antifungal rate/% | 1 | -0.927** | -0.952** | -0.998** | -0.985** | 0.966** |
| SOD活力SOD activity/(U·g-1) | -0.927** | 1 | 0.811* | 0.902* | 0.857* | 0.842* |
| POD活力POD activity/(U·g-1) | -0.952** | 0.811* | 1 | 0.963** | 0.945** | 0.892* |
| CAT活力CAT activity/(U·g-1) | -0.998** | 0.902* | 0.963** | 1 | 0.992** | 0.971** |
| 可溶性蛋白含量 Soluble protein content /(mg·g-1) | -0.985** | 0.857* | 0.945** | 0.992** | 1 | 0.991** |
| 可溶性糖含量 Soluble sugar content /(mg·g-1) | -0.966** | 0.842* | 0.892* | 0.971** | 0.991** | 1 |
| 平均值Mean | -0.966 | 0.868 | 0.913 | 0.965 | 0.954 | 0.932 |
Table 1 Correlation coefficient among indexes
| 指标 Index | 抑菌率 Antifungal rate/% | SOD活力 SOD activity/ (U·g-1) | POD活力 POD activity/ (U·g-1) | CAT活力 CAT activity/ (U·g-1) | 可溶性蛋白含量 Soluble protein content/ (mg·g-1) | 可溶性糖含量 Soluble sugar content/ (mg·g-1) |
|---|---|---|---|---|---|---|
| 抑菌率Antifungal rate/% | 1 | -0.927** | -0.952** | -0.998** | -0.985** | 0.966** |
| SOD活力SOD activity/(U·g-1) | -0.927** | 1 | 0.811* | 0.902* | 0.857* | 0.842* |
| POD活力POD activity/(U·g-1) | -0.952** | 0.811* | 1 | 0.963** | 0.945** | 0.892* |
| CAT活力CAT activity/(U·g-1) | -0.998** | 0.902* | 0.963** | 1 | 0.992** | 0.971** |
| 可溶性蛋白含量 Soluble protein content /(mg·g-1) | -0.985** | 0.857* | 0.945** | 0.992** | 1 | 0.991** |
| 可溶性糖含量 Soluble sugar content /(mg·g-1) | -0.966** | 0.842* | 0.892* | 0.971** | 0.991** | 1 |
| 平均值Mean | -0.966 | 0.868 | 0.913 | 0.965 | 0.954 | 0.932 |
| 样品 Sample | 原始序列数 Raw reads | 质控后序列数 Clean reads | 质控后序列比例 Clean reads rate/% | 错误率 Error rate/% | Q20/% | Q30/% | GC 含量 GC content/% | 覆盖率 Mapped ratios/% |
|---|---|---|---|---|---|---|---|---|
| C1 | 44 046 600 | 42 603 392 | 96.72 | 0.03 | 97.00 | 91.69 | 52.60 | 78.75 |
| C2 | 43 260 998 | 41 992 094 | 97.07 | 0.03 | 96.99 | 91.62 | 52.87 | 79.61 |
| C3 | 58 845 594 | 56 937 404 | 96.76 | 0.03 | 98.03 | 94.15 | 52.30 | 79.27 |
| T1 | 54 973 812 | 52 895 734 | 96.22 | 0.03 | 97.93 | 93.83 | 52.31 | 78.44 |
| T2 | 58 068 196 | 55 878 726 | 96.23 | 0.03 | 97.95 | 93.99 | 52.27 | 78.47 |
| T3 | 57 570 042 | 55 363 728 | 96.17 | 0.03 | 97.84 | 93.66 | 52.55 | 79.33 |
| 综合Total | 316 765 242 | 305 671 078 | 96.50 |
Table 2 Quality control data statistics
| 样品 Sample | 原始序列数 Raw reads | 质控后序列数 Clean reads | 质控后序列比例 Clean reads rate/% | 错误率 Error rate/% | Q20/% | Q30/% | GC 含量 GC content/% | 覆盖率 Mapped ratios/% |
|---|---|---|---|---|---|---|---|---|
| C1 | 44 046 600 | 42 603 392 | 96.72 | 0.03 | 97.00 | 91.69 | 52.60 | 78.75 |
| C2 | 43 260 998 | 41 992 094 | 97.07 | 0.03 | 96.99 | 91.62 | 52.87 | 79.61 |
| C3 | 58 845 594 | 56 937 404 | 96.76 | 0.03 | 98.03 | 94.15 | 52.30 | 79.27 |
| T1 | 54 973 812 | 52 895 734 | 96.22 | 0.03 | 97.93 | 93.83 | 52.31 | 78.44 |
| T2 | 58 068 196 | 55 878 726 | 96.23 | 0.03 | 97.95 | 93.99 | 52.27 | 78.47 |
| T3 | 57 570 042 | 55 363 728 | 96.17 | 0.03 | 97.84 | 93.66 | 52.55 | 79.33 |
| 综合Total | 316 765 242 | 305 671 078 | 96.50 |
| 二级类别 Second category | 通路名称 Pathway name | 基因编号 Gene ID | 基因名称 Gene name | Log2FC | P-value | 上下调Regulation |
|---|---|---|---|---|---|---|
| 碳水化合物代谢Carbohydrate metabolism | 半乳糖代谢 Galactose metabolism | FGSG_03569 | Galactose oxidase precursor | 1.20 | 3.85×10-3 | Up |
| FGSG_11032 | Galactose oxidase precursor | 2.11 | 3.43×10-3 | Up | ||
| FGSG_00096 | Hypothetical protein | 2.57 | 3.88×10-8 | Up | ||
| FGSG_03288 | Hypothetical protein | 1.77 | 8.10×10-4 | Up | ||
| FGSG_08415 | Hypothetical protein | 1.90 | 1.13×10-7 | Up | ||
| FGSG_13188 | Hypothetical protein | -1.78 | 2.89×10-3 | Down | ||
| 氨基糖和核苷糖代谢途径 Amino sugar and nucleotide sugar metabolism | FGSG_10327 | Chitin synthase 3 | -1.22 | 5.28×10-6 | Down | |
| FGSG_03591 | 42 kD endochitinase precursor | -1.77 | 1.93×10-3 | Down | ||
| FGSG_08773 | Mannose-1-phosphate guanyltransferase | 1.19 | 3.13×10-3 | Up | ||
| FGSG_04752 | Hypothetical protein | 6.07 | 5.10×10-5 | Up | ||
| FGSG_02355 | Hypothetical protein | -1.69 | 1.39×10-3 | Down | ||
| FGSG_04903 | Hypothetical protein | 1.88 | 3.10×10-3 | Up | ||
| FGSG_13188 | Hypothetical protein | -1.78 | 2.89×10-3 | Down | ||
| FGSG_13883 | Hypothetical protein | -1.05 | 9.45×10-4 | Down | ||
| 脂代谢 Lipid metabolism | 类角甾醇生物合成Steroid biosynthesis | FGSG_02502 | C-5 sterol desaturase | 3.39 | 2.14×10-10 | Up |
| FGSG_02346 | Delta(14)-sterol reductase | 1.10 | 3.88×10-3 | Up | ||
| FGSG_09830 | C-4 methylsterol oxidase | 1.46 | 4.18×10-3 | Up | ||
| FGSG_02016 | Hypothetical protein | -1.32 | 1.08×10-4 | Down | ||
| FGSG_06215 | Hypothetical protein | 1.38 | 3.87×10-3 | Up | ||
| 氨基酸代谢Amino acid metabolism | 缬氨酸、亮氨酸和异亮氨酸生物合成 Valine, leucine and isoleucine biosynthesis | FGSG_09589 | 3-isopropylmalate dehydratase | -1.05 | 4.61×10-3 | Down |
| FGSG_10118 | Ketol-acid reductoisomerase | 1.17 | 1.28×10-3 | Up | ||
| FGSG_10119 | Hypothetical protein | 1.33 | 1.07×10-6 | Up | ||
| FGSG_04570 | Hypothetical protein | -1.52 | 8.46×10-5 | Down | ||
| 谷胱甘肽代谢 Glutathione metabolism | FGSG_05903 | Ornithine decarboxylase | -1.36 | 1.82×10-7 | Down | |
| FGSG_00172 | Hypothetical protein | -1.47 | 3.62×10-3 | Down | ||
| FGSG_00443 | Hypothetical protein | -1.19 | 2.42×10-4 | Down | ||
| FGSG_12154 | Hypothetical protein | 1.58 | 7.48×10-6 | Up | ||
| FGSG_04902 | Hypothetical protein | 4.34 | 3.65×10-4 | Up | ||
| 辅助因子和维生素代谢Metabolism of cofactors and vitamins | 泛酸盐和乙酰辅酶A生物合成 Pantothenate and CoA biosynthesis | FGSG_10118 | Ketol-acid reductoisomeras | 1.17 | 1.28×10-3 | Up |
| FGSG_12108 | Hypothetical protein | 1.27 | 3.02×10-3 | Up | ||
| FGSG_04570 | Hypothetical protein | 1.52 | 8.46×10-5 | Uup | ||
| FGSG_03176 | Hypothetical protein | -1.21 | 1.17×10-3 | Down |
Table 3 Significantly enriched of differentially expressed genes and pathways by KEGG
| 二级类别 Second category | 通路名称 Pathway name | 基因编号 Gene ID | 基因名称 Gene name | Log2FC | P-value | 上下调Regulation |
|---|---|---|---|---|---|---|
| 碳水化合物代谢Carbohydrate metabolism | 半乳糖代谢 Galactose metabolism | FGSG_03569 | Galactose oxidase precursor | 1.20 | 3.85×10-3 | Up |
| FGSG_11032 | Galactose oxidase precursor | 2.11 | 3.43×10-3 | Up | ||
| FGSG_00096 | Hypothetical protein | 2.57 | 3.88×10-8 | Up | ||
| FGSG_03288 | Hypothetical protein | 1.77 | 8.10×10-4 | Up | ||
| FGSG_08415 | Hypothetical protein | 1.90 | 1.13×10-7 | Up | ||
| FGSG_13188 | Hypothetical protein | -1.78 | 2.89×10-3 | Down | ||
| 氨基糖和核苷糖代谢途径 Amino sugar and nucleotide sugar metabolism | FGSG_10327 | Chitin synthase 3 | -1.22 | 5.28×10-6 | Down | |
| FGSG_03591 | 42 kD endochitinase precursor | -1.77 | 1.93×10-3 | Down | ||
| FGSG_08773 | Mannose-1-phosphate guanyltransferase | 1.19 | 3.13×10-3 | Up | ||
| FGSG_04752 | Hypothetical protein | 6.07 | 5.10×10-5 | Up | ||
| FGSG_02355 | Hypothetical protein | -1.69 | 1.39×10-3 | Down | ||
| FGSG_04903 | Hypothetical protein | 1.88 | 3.10×10-3 | Up | ||
| FGSG_13188 | Hypothetical protein | -1.78 | 2.89×10-3 | Down | ||
| FGSG_13883 | Hypothetical protein | -1.05 | 9.45×10-4 | Down | ||
| 脂代谢 Lipid metabolism | 类角甾醇生物合成Steroid biosynthesis | FGSG_02502 | C-5 sterol desaturase | 3.39 | 2.14×10-10 | Up |
| FGSG_02346 | Delta(14)-sterol reductase | 1.10 | 3.88×10-3 | Up | ||
| FGSG_09830 | C-4 methylsterol oxidase | 1.46 | 4.18×10-3 | Up | ||
| FGSG_02016 | Hypothetical protein | -1.32 | 1.08×10-4 | Down | ||
| FGSG_06215 | Hypothetical protein | 1.38 | 3.87×10-3 | Up | ||
| 氨基酸代谢Amino acid metabolism | 缬氨酸、亮氨酸和异亮氨酸生物合成 Valine, leucine and isoleucine biosynthesis | FGSG_09589 | 3-isopropylmalate dehydratase | -1.05 | 4.61×10-3 | Down |
| FGSG_10118 | Ketol-acid reductoisomerase | 1.17 | 1.28×10-3 | Up | ||
| FGSG_10119 | Hypothetical protein | 1.33 | 1.07×10-6 | Up | ||
| FGSG_04570 | Hypothetical protein | -1.52 | 8.46×10-5 | Down | ||
| 谷胱甘肽代谢 Glutathione metabolism | FGSG_05903 | Ornithine decarboxylase | -1.36 | 1.82×10-7 | Down | |
| FGSG_00172 | Hypothetical protein | -1.47 | 3.62×10-3 | Down | ||
| FGSG_00443 | Hypothetical protein | -1.19 | 2.42×10-4 | Down | ||
| FGSG_12154 | Hypothetical protein | 1.58 | 7.48×10-6 | Up | ||
| FGSG_04902 | Hypothetical protein | 4.34 | 3.65×10-4 | Up | ||
| 辅助因子和维生素代谢Metabolism of cofactors and vitamins | 泛酸盐和乙酰辅酶A生物合成 Pantothenate and CoA biosynthesis | FGSG_10118 | Ketol-acid reductoisomeras | 1.17 | 1.28×10-3 | Up |
| FGSG_12108 | Hypothetical protein | 1.27 | 3.02×10-3 | Up | ||
| FGSG_04570 | Hypothetical protein | 1.52 | 8.46×10-5 | Uup | ||
| FGSG_03176 | Hypothetical protein | -1.21 | 1.17×10-3 | Down |
| [1] |
Fonseca-Guerra IR, Chiquillo-Pompeyo JC, Benavides Rozo ME, Díaz Ovalle JF. Fusarium spp. associated with Chenopodium quinoa crops in Colombia[J]. Sci Rep, 2022, 12(1): 20841.
doi: 10.1038/s41598-022-24908-w pmid: 36460698 |
| [2] |
Sanogo S, Zhang JF. Resistance sources, resistance screening techniques and disease management for Fusarium wilt in cotton[J]. Euphytica, 2016, 207(2): 255-271.
doi: 10.1007/s10681-015-1532-y URL |
| [3] |
Tian DD, Song XP, Li CS, et al. Antifungal mechanism of Bacillus amyloliquefaciens strain GKT04 against Fusarium wilt revealed using genomic and transcriptomic analyses[J]. MicrobiologyOpen, 2021, 10(3): e1192.
doi: 10.1002/mbo3.v10.3 URL |
| [4] | 张瑶, 高弢, 马桂珍, 等. 基于转录组测序技术分析愈创木酚对禾谷镰刀菌的抑菌机制[J]. 江苏农业学报, 2022, 38(2): 343-351. |
| Zhang Y, Gao T, Ma GZ, et al. Analysis on the antifungal mechanism of Fusarium graminearum treated by guaiacol based on transcriptome sequencing[J]. Jiangsu J Agric Sci, 2022, 38(2): 343-351. | |
| [5] | 付显惠, 沈启芳, 黄盼盼, 等. 禾谷镰刀菌组蛋白乙酰转移酶FgHat1在DON毒素生物合成中的功能[J]. 植物病理学报, 2022, 52(2): 145-155. |
| Fu XH, Shen QF, Huang PP, et al. The histone acetyltransferase FgHat1 is important for DON biosynthesis in Fusarium graminearum[J]. Acta Phytopathol Sin, 2022, 52(2): 145-155. | |
| [6] |
Zhu ZH, Zhao SJ, Wang CH. β-carboline alkaloids from Peganum harmala inhibit Fusarium oxysporum from Codonopsis radix through damaging the cell membrane and inducing ROS accumulation[J]. Pathogens, 2022, 11(11): 1341.
doi: 10.3390/pathogens11111341 URL |
| [7] | 罗晓娇, 孙静, 陆颖健. 解淀粉芽孢杆菌中脂肽的生物合成、抑菌机理及应用的研究进展[J]. 食品工业科技, 2022, 43(19): 462-470. |
| Luo XJ, Sun J, Lu YJ. Research progress in the biosynthesis, antimicrobial mechanism, and application of lipopeptides in Bacillus amyloliquefaciens[J]. Sci Technol Food Ind, 2022, 43(19): 462-470. | |
| [8] | 金清, 肖明. 新型抗菌肽——表面活性素、伊枯草菌素和丰原素[J]. 微生物与感染, 2018, 13(1): 56-64. |
| Jin Q, Xiao M. Novel antimicrobial peptides: surfactin, iturin and fengycin[J]. J Microbes Infect, 2018, 13(1): 56-64. | |
| [9] |
Li SJ, Xu XG, Zhao TY, et al. Screening of Bacillus velezensis E2 and the inhibitory effect of its antifungal substances on Aspergillus flavus[J]. Foods, 2022, 11(2): 140.
doi: 10.3390/foods11020140 URL |
| [10] |
Saad A, Christopher J, Martin A, et al. Fusarium pseudogramine-arum and F. culmorum affect the root system architecture of bread wheat[J]. Crop J, 2023, 11(1): 316-321.
doi: 10.1016/j.cj.2022.08.013 URL |
| [11] | 高景凯, 赵前程, 孙佳琦, 等. 广东石豆兰内生细菌分离鉴定及抑菌活性[J]. 安徽科技学院学报, 2019, 33(3): 15-21. |
| Gao JK, Zhao QC, Sun JQ, et al. Isolation and identification of endophytic bacteria in Bulbophyllum kwangtungense schltr for antimicrobial activities[J]. J Anhui Sci Technol Univ, 2019, 33(3): 15-21. | |
| [12] |
张祥丽, 曹瑱艳, 杨怡华, 等. 铁皮石斛镰刀菌根腐病病原菌的鉴定及其对链霉菌发酵液的敏感性分析[J]. 中国生物防治学报, 2022, 38(1): 258-266.
doi: 10.16409/j.cnki.2095-039x.2022.02.004 |
| Zhang XL, Cao TY, Yang YH, et al. Identification of the pathogen causing root rot of Dendrobium officinale and sensitivity to the fermentation broth of Streptomyces[J]. Chin J Biol Contr, 2022, 38(1): 258-266. | |
| [13] | 苗永美, 韩朔, 苗翠苹, 等. 石豆兰内生细菌分离鉴定、发酵条件优化及对棉花枯萎病菌的抑制[J]. 天然产物研究与开发, 2022, 34(4): 656-664. |
| Miao YM, Han S, Miao CP, et al. Isolation, identification and fermentation conditions optimization of endophytes from Bulbophyllum sp. and its inhibitory effect on Fusarium oxysporum f. sp. vasinfectum[J]. Nat Prod Res Dev, 2022, 34(4): 656-664. | |
| [14] |
Wu SM, Liu G, Zhou SN, et al. Characterization of antifungal lipopeptide biosurfactants produced by marine bacterium Bacillus sp. CS30[J]. Mar Drugs, 2019, 17(4): 199.
doi: 10.3390/md17040199 URL |
| [15] | 韩占红. 扩展青霉麦角甾醇合成相关基因erg4A、erg4B和erg4C的生物学功能研究[D]. 兰州: 甘肃农业大学, 2021. |
| Han ZH. Biological function study on ergosterol synthesis-related genes erg4A, erg4B and erg4C of Penicillium expansum[D]. Lanzhou: Gansu Agricultural University, 2021. | |
| [16] | 杨晓燕, 王钰鑫, 叶伟伟, 等. 枯草芽孢杆菌对几种植物病原真菌的抑菌活性[J]. 工业微生物, 2018, 48(6): 32-38. |
| Yang XY, Wang YX, Ye WW, et al. Antifungal activity of Bacillus subtilis strain against several plant pathogenic fungi[J]. Ind Microbiol, 2018, 48(6): 32-38. | |
| [17] | 邵林, 李璟, 乐露, 等. 枯草芽孢杆菌和沼泽红假单胞菌对鲫鱼免疫机能的影响及其代谢产物的抑菌活性研究[J]. 黑龙江畜牧兽医, 2018(20): 176-178. |
| Shao L, Li J, Le L, et al. The effect of Bacillus subtilis and Rhodopseudomonas palustris on the immune function of Carassius auratus and their antimicrobial activity[J]. Heilongjiang Anim Sci Vet Med, 2018(20): 176-178. | |
| [18] | 李颖, 金鑫, 沙海天, 等. 枯草芽孢杆菌SH-1发酵条件优化及抑菌活性物质稳定性研究[J]. 食品科技, 2016, 41(9): 25-29. |
|
Li Y, Jin X, Sha HT, et al. Optimization of fermentation conditions of Bacillus subtilis SH-1 and stability of antimicrobial substances[J]. Food Sci Technol, 2016, 41(9): 25-29.
doi: 10.1590/fst.27119 URL |
|
| [19] | 吴越, 王世锋, 陈鑫, 等. 枯草芽孢杆菌HAINUP40的培养条件及其抑菌物质分析[J]. 基因组学与应用生物学, 2020, 39(6): 2638-2645. |
| Wu Y, Wang SF, Chen X, et al. Culture conditions and analysis of bacteriostatic substances of Bacillus subtillis HAINUP40[J]. Genom Appl Biol, 2020, 39(6): 2638-2645. | |
| [20] | 邓阳, 董伟洁, 朱永明, 等. 枯草芽孢杆菌J-4菌株胞外产抑菌物质性质的初探[J]. 黑龙江畜牧兽医, 2018(7): 4-8, 244. |
| Deng Y, Dong WJ, Zhu YM, et al. Preliminary study on properties of extracellular antibacterial substances from Bacillus subtilis strain J-4[J]. Heilongjiang Anim Sci Vet Med, 2018(7): 4-8, 244. | |
| [21] | 李丽, 程煜, 袁建琴. 枯草芽孢杆菌S6抗棉花枯萎病菌拮抗蛋白的分离纯化与抑菌活性研究[J]. 天津农业科学, 2020, 26(6): 3-6. |
| Li L, Cheng Y, Yuan JQ. Purification and antibacterial activity of antagonistic protein against bacterial Fusarium wilt from Bacillus subtilis S6[J]. Tianjin Agric Sci, 2020, 26(6): 3-6. | |
| [22] | 曾国洪, 丛丽娜, 毕楠. 枯草芽孢杆菌诱变株产抗菌脂肽的特性[J]. 大连工业大学学报, 2019, 38(4): 235-238. |
| Zeng GH, Cong LN, Bi N. Characteristics of antimicrobial lipopeptides produced by Bacillus subtilis mutant strains[J]. J Dalian Polytech Univ, 2019, 38(4): 235-238. | |
| [23] | 梁艳琼, 唐文, 董文敏, 等. 枯草芽孢杆菌菌株Czk1挥发性物质的抑菌活性及其组分分析[J]. 南方农业学报, 2019, 50(11): 2465-2474. |
| Liang YQ, Tang W, Dong WM, et al. Antifugal effect and components analysis of volatile organic compounds from Bacillus subtilis Czk1[J]. J South Agric, 2019, 50(11): 2465-2474. | |
| [24] | 桑建伟, 杨扬, 陈奕鹏, 等. 内生解淀粉芽孢杆菌BEB17脂肽类和聚酮类化合物的抑菌活性分析[J]. 植物病理学报, 2018, 48(3): 402-412. |
| Sang JW, Yang Y, Chen YP, et al. Antibacterial activity analysis of lipopeptide and polyketide compounds produced by endophytic bacteria Bacillus amyloliquefaciens BEB17[J]. Acta Phytopathol Sin, 2018, 48(3): 402-412. | |
| [25] |
Kurtz MB, Heath IB, Marrinan J, et al. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against(1, 3)-beta-D-glucan synthase[J]. Antimicrob Agents Chemother, 1994, 38(7): 1480-1489.
doi: 10.1128/AAC.38.7.1480 pmid: 7979276 |
| [26] | 崔旭. D-半乳糖衰老模型及其氧化损伤机理与α-硫辛酸的干预作用研究[D]. 北京: 中国协和医科大学, 2004. |
| Cui X. Studies on D-galactose aging model and its mechanism of oxidative damage and antagonistic effects of α-lipoic acid[D]. Beijing: Peking Union Medical College, 2004. | |
| [27] | 宋修仕. 禾谷镰刀菌细胞壁形成相关基因及其 RNAi 片段功能鉴定[D]. 武汉: 华中农业大学, 2015. |
| Song XS. Functional identification of cell wall biosynthesis-related genes and their RNAi molecules in Fusarium graminearum[D]. Wuhan: Huazhong Agricultural University, 2015. | |
| [28] | 刘向勇, 张小华, 王跃嗣. 酿酒酵母CHS3基因缺失对热耐受性的影响[J]. 中国酿造, 2010, 29(5): 127-129. |
| Liu XY, Zhang XH, Wang YS. Effect of deletion of CHS3 gene on the thermotolerance in Saccharomyces cerevisiae[J]. China Brew, 2010, 29(5): 127-129. | |
| [29] |
Zhou L, Song CX, Muñoz CY, et al. Bacillus cabrialesii BH5 protects tomato plants against Botrytis cinerea by production of specific antifungal compounds[J]. Front Microbiol, 2021, 12: 707609.
doi: 10.3389/fmicb.2021.707609 URL |
| [30] | 方欣, 王菲菲, 范中玥, 等. FgLEU1在禾谷镰刀菌亮氨酸生物合成及产毒致病中起关键作用[J]. 植物保护学报, 2022, 49(2): 497-507. |
| Fang X, Wang FF, Fan ZY, et al. FgLEU1 is crucial for leucine biosynthesis, virulence and DON production in fungal pathogen Fusarium graminearum[J]. J Plant Prot, 2022, 49(2): 497-507. |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [3] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [4] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [5] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [6] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [7] | WANG Wei-chen, ZHAO Jin, HUANG Wei-yi, GUO Xin-zhu, LI Wan-ying, ZHANG Zhuo. Research Progress in Metabolites Produced by Bacillus Against Three Common Plant Pathogenic Fungi [J]. Biotechnology Bulletin, 2023, 39(3): 59-68. |
| [8] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [9] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| [10] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [11] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| [12] | XU Jin, LI Tao, LI Chu-lin, ZHU Shun-ni, WANG Zhong-ming, XIANG Wen-zhou. Effects of Temperature on the Growth,Total Lipid and Eicosapentaenoic Acid Synthesis of Eustigmatos sp. [J]. Biotechnology Bulletin, 2022, 38(6): 261-271. |
| [13] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [14] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [15] | GUAN Yi, LI Xin, WANG Ding-yi, DU Xi, ZHANG Long-bin, YE Xiu-yun. Functional Study of BbRho5 on the Growth Rate of Beauveria bassiana [J]. Biotechnology Bulletin, 2022, 38(2): 132-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||