








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 118-128.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0177
Previous Articles Next Articles
XING Li-nan( ), ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen(
), ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen( )
)
Received:2024-02-23
Online:2024-08-26
Published:2024-06-26
Contact:
HUO Xiu-wen
E-mail:1760349574@qq.com;huoxiuwen@imau.edu.cn
XING Li-nan, ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen. Analysis of DoWRKY40 Gene Expression Characteristics and Screening of Interacting Proteins in Yam[J]. Biotechnology Bulletin, 2024, 40(8): 118-128.
| 引物名称Primer name | 引物序列 Primer sequences(5'-3') | 引物用途Primer usage |
|---|---|---|
| WRKY40-F | TTCTTCTTCTTCTTCTTCTTCT | 已知序列验证Sequence verification |
| WRKY40-R | AACATTCAAAAGTCTCCCCCA | |
| WRKY40-qF | AATGGAGCAATCATCACTTA | RT-qPCR |
| WRKY40-qR | AACATCTCACTCAATCTTTC | |
| 18S-F | CCATAAACGATGCCGACCAG | 18S rRNA |
| 18S-R | AGCCTTGCGACCATACTCCC | |
| WRKY40-SF | CGGGGTACCATGGAATCAATAACAATGGAGC | 亚细胞定位Subcellular localization |
| WRKY40-SR | GCGGATCCTATTTGGAGAAAAACTAAGAA | |
| WRKY40-Y-F | GCGGATCCATGGAATCAATAACAATGGAGC | 诱饵载体构建Decoy vector construction |
| WRKY40-Y-R | CTGCAGATTTGGAGAAAAACTAAGAATTTTT | |
| 3'AD | GTGAACTTGCGGGGTTTTTCAG | 酵母质粒PCR Yeast plasmid PCR |
| 5'AD | CTATTCGATGATGAAGATACCCC |
Table 1 Primer sequences related to WRKY40
| 引物名称Primer name | 引物序列 Primer sequences(5'-3') | 引物用途Primer usage |
|---|---|---|
| WRKY40-F | TTCTTCTTCTTCTTCTTCTTCT | 已知序列验证Sequence verification |
| WRKY40-R | AACATTCAAAAGTCTCCCCCA | |
| WRKY40-qF | AATGGAGCAATCATCACTTA | RT-qPCR |
| WRKY40-qR | AACATCTCACTCAATCTTTC | |
| 18S-F | CCATAAACGATGCCGACCAG | 18S rRNA |
| 18S-R | AGCCTTGCGACCATACTCCC | |
| WRKY40-SF | CGGGGTACCATGGAATCAATAACAATGGAGC | 亚细胞定位Subcellular localization |
| WRKY40-SR | GCGGATCCTATTTGGAGAAAAACTAAGAA | |
| WRKY40-Y-F | GCGGATCCATGGAATCAATAACAATGGAGC | 诱饵载体构建Decoy vector construction |
| WRKY40-Y-R | CTGCAGATTTGGAGAAAAACTAAGAATTTTT | |
| 3'AD | GTGAACTTGCGGGGTTTTTCAG | 酵母质粒PCR Yeast plasmid PCR |
| 5'AD | CTATTCGATGATGAAGATACCCC |
| 软件 Software | 用途 Usage |
|---|---|
| NCBI ORF Finder | CDS序列分析 CDS sequence analysis |
| Prot Param | 蛋白理化性质分析Analysis in physicochemical properties of coding protein |
| TMHMM Server 2.0 | 跨膜结构域预测 Prediction of protein transmembrane domain |
| SOPMA | 蛋白结构预测 Prediction of protein structure |
| DNAMAN | 氨基酸序列比对 Alignment of amino acid sequences |
| CDD | 保守结构域分析 Analysis of conservative domain |
| NCBI Blast | 同源性搜索及比对Homology search and alignment of amino acid sequence |
| MEGA 5.0 | 系统进化树的构建 Construction of phylogenetic tree |
Table 2 Related software for DoWRKY40 bioinformatics analysis
| 软件 Software | 用途 Usage |
|---|---|
| NCBI ORF Finder | CDS序列分析 CDS sequence analysis |
| Prot Param | 蛋白理化性质分析Analysis in physicochemical properties of coding protein |
| TMHMM Server 2.0 | 跨膜结构域预测 Prediction of protein transmembrane domain |
| SOPMA | 蛋白结构预测 Prediction of protein structure |
| DNAMAN | 氨基酸序列比对 Alignment of amino acid sequences |
| CDD | 保守结构域分析 Analysis of conservative domain |
| NCBI Blast | 同源性搜索及比对Homology search and alignment of amino acid sequence |
| MEGA 5.0 | 系统进化树的构建 Construction of phylogenetic tree |
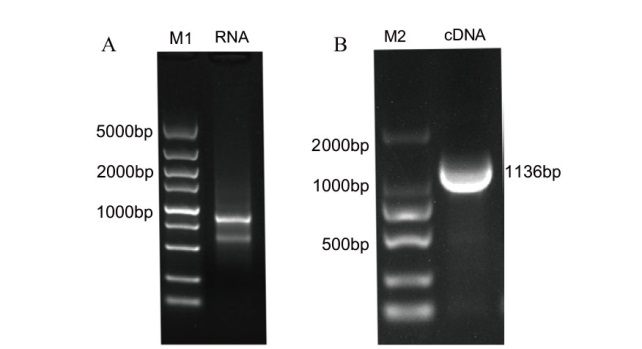
Fig. 1 Extraction of RNA of yam and PCR of DoWRKY40 gene A: Photo of RNA electrophoresis detection; B: Verification of known DoWRKY40 sequences; M1: DL5000; M2: DL2000

Fig. 2 Coding sequence and deduced amino acid sequence of DoWRKY40 Red box: WRKY protein conserved sequence WRKYGQK; underline: C2H2 domain; *: termination codon
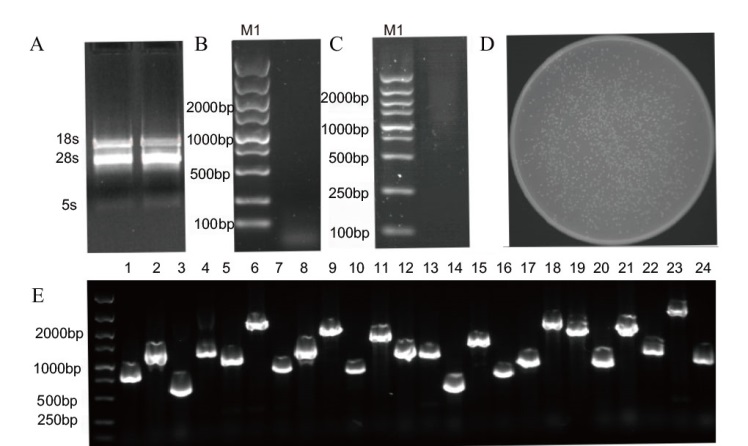
Fig. 7 Construction of cDNA library and identification A: Photo of RNA electrophoresis detection. B: Photo of ds cDNA electrophoresis detection. C: Photo of homogenization and defragmentation electrophoresis detection. Escherichia coli colony count (D) and PCR identification of 24 monoclonal insertion fragments (E); M1: DL5000 marker. 1-24: Selected 24 cloned colonies PCR
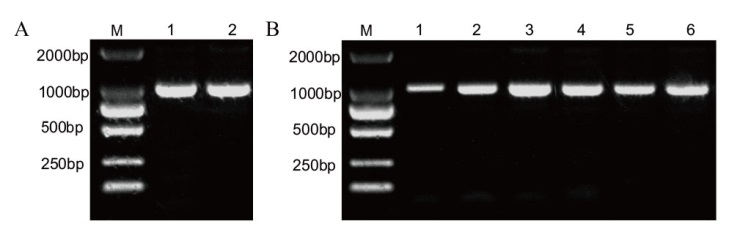
Fig. 8 Construction of bait vector and PCR identification of colony A: PCR of DoWRKY40 plasmid; 1-2: PCR amplification products; B: PCR of pGBKT7-DoWRKY40 colony; 1-6: PCR results of pGBKT7-DoWRKY40 positive clonal colony; M: DL5000 DNA marker
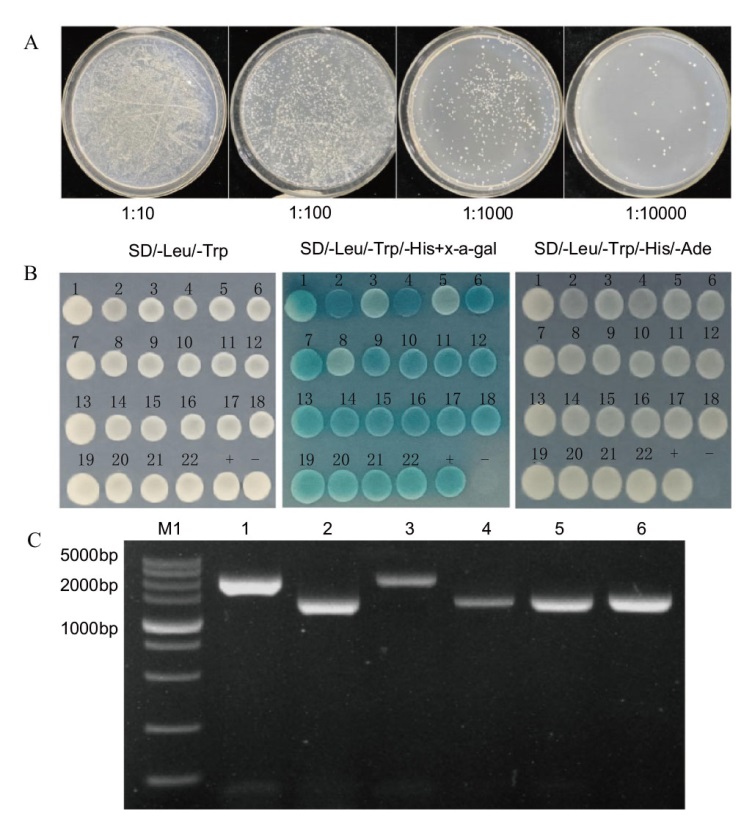
Fig. 10 Efficiency of cDNA libary screening (A), DoWRKY40 interaction protein screening (B) and plasmid PCR detection (C) B: 1-22: Numbering of screened positive clones; +: positive control; -: negative control; C: M1: DL5000 DNA marker; 1-6: PCR results of 1-6 plasmid
| 序号 No. | NCBI比对结果 NCBI blast results | E-value | 相似度 Similarity/% | 登录号 Accession number | 克隆数 Clone count |
|---|---|---|---|---|---|
| 1 | 转录因子EGL1 Transcription factor EGL1 | 0.0 | 93 | XM-039282616.1 | 1 |
| 2 | DNA结合蛋白BIN4 DNA-binding protein BIN4 | 0.0 | 94 | XM-039264312.1 | 3 |
| 3 | 蛋白磷酸酶2C Protein phosphatase 2C | 0.0 | 86 | XM-039277539.1 | 1 |
| 4 | BAG家族分子伴侣调控因子7 BAG family molecular chaperone regulator 7-like | 0.0 | 95 | XM-039266610.1 | 1 |
Table 3 BLAST analysis of candidate proteins interacted with DoWRKY40
| 序号 No. | NCBI比对结果 NCBI blast results | E-value | 相似度 Similarity/% | 登录号 Accession number | 克隆数 Clone count |
|---|---|---|---|---|---|
| 1 | 转录因子EGL1 Transcription factor EGL1 | 0.0 | 93 | XM-039282616.1 | 1 |
| 2 | DNA结合蛋白BIN4 DNA-binding protein BIN4 | 0.0 | 94 | XM-039264312.1 | 3 |
| 3 | 蛋白磷酸酶2C Protein phosphatase 2C | 0.0 | 86 | XM-039277539.1 | 1 |
| 4 | BAG家族分子伴侣调控因子7 BAG family molecular chaperone regulator 7-like | 0.0 | 95 | XM-039266610.1 | 1 |
| [1] | Shan N, Wang PT, Zhu QL, et al. Comprehensive characterization of yam tuber nutrition and medicinal quality of Dioscorea opposita and D. alata from different geographic groups in China[J]. J Integr Agric, 2020, 19(11): 2839-2848. |
| [2] | Cao TX, Sun JY, Shan N, et al. Uncovering the genetic diversity of yams(Dioscorea spp.)in China by combining phenotypic trait and molecular marker analyses[J]. Ecol Evol, 2021, 11(15): 9970-9986. |
| [3] | 陈妍, 张艳芳, 赵令敏, 等. 光照强度对山药光合特性的影响和Rubisco羧化酶基因的克隆及表达分析[J]. 植物生理学报, 2023, 59(6): 1169-1183. |
| Chen Y, Zhang YF, Zhao LM, et al. Effects of light intensity on photosynthetic characteristics of yam(Dioscorea opposita)and cloning and expression analysis of Rubisco carboxylase gene[J]. Plant Physiol J, 2023, 59(6): 1169-1183. | |
| [4] | 葛明然, 张艳芳, 邢丽南, 等. 山药(Dioscorea opposita Thunb.)赤霉素合成酶基因DoGA20ox1的克隆及功能研究[J]. 西北农业学报, 2023, 32(6): 919-928. |
| Ge MR, Zhang YF, Xing LN, et al. Cloning and function of gibberell in synthase gene DoGA20ox1 in yam(Dioscorea opposita Thunb.)[J]. Acta Agric Boreali Occidentalis Sin, 2023, 32(6): 919-928. | |
| [5] | Zhu H, Zhou YY, Zhai H, et al. A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis[J]. Biomolecules, 2020, 10(4): 506. |
| [6] | Jang JY, Choi CH, Hwang DJ. The WRKY superfamily of rice transcription factors[J]. Plant Pathol J, 2010, 26(2): 110-114. |
| [7] | McGregor C. Differential expression and detection of transcripts in sweetpotato(Ipomoea batatas(L.)Lam.)using cDNA microarrays[D]. Baton Rouge, USA: Louisiana State University Libraries, 2006. |
| [8] | Yu YC, Hu RB, Wang HM, et al. MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering[J]. Plant Sci, 2013, 212: 1-9. |
| [9] | 陈凤颖, 龙小琴, 聂聪, 等. 转录因子WRKY71对拟南芥根系发育的影响[J]. 植物生理学报, 2022, 58(2): 363-370. |
| Chen FY, Long XQ, Nie C, et al. Effects of transcription factor WRKY71 on root development of Arabidopsis thaliana[J]. Plant Physiol J, 2022, 58(2): 363-370. | |
| [10] | Huang Y, Feng CZ, Ye Q, et al. Correction: Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development[J]. PLoS Genet, 2019, 15(3): e1008032. |
| [11] | 田媛, 郑锦城. 刺梨WRKY基因家族鉴定及其在不同组织中的表达分析[J]. 分子植物育种, 2024, 22(4): 1075-1085. |
| Tian Y, Zheng JC. Identification and expression analysis in different tissues of WRKY transcription factors in Rosa roxburghii tratt[J]. Mol Plant Breed, 2024, 22(4): 1075-1085. | |
| [12] |
Wang DJ, Wang L, Su WH, et al. A class III WRKY transcription factor in sugarcane was involved in biotic and abiotic stress responses[J]. Sci Rep, 2020, 10(1): 20964.
doi: 10.1038/s41598-020-78007-9 pmid: 33262418 |
| [13] | Rosado D, Ackermann A, Spassibojko O, et al. WRKY transcription factors and ethylene signaling modify root growth during the shade-avoidance response[J]. Plant Physiol, 2022, 188(2): 1294-1311. |
| [14] | Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato[J]. Mol Gen Genet, 1994, 244(6): 563-571. |
| [15] |
Eulgem T, Rushton PJ, Robatzek S, et al. The WRKY superfamily of plant transcription factors[J]. Trends Plant Sci, 2000, 5(5): 199-206.
doi: 10.1016/s1360-1385(00)01600-9 pmid: 10785665 |
| [16] | Wei W, Hu Y, Han YT, et al. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses[J]. Plant Physiol Biochem, 2016, 105: 129-144. |
| [17] | Jing ZB, Liu ZD. Genome-wide identification of WRKY transcription factors in kiwifruit(Actinidia spp.)and analysis of WRKY expression in responses to biotic and abiotic stresses[J]. Genes Genomics, 2018, 40(4): 429-446. |
| [18] | Wu KL, Guo ZJ, Wang HH, et al. The WRKY family of transcription factors in rice and Arabidopsis and their origins[J]. DNA Res, 2005, 12(1): 9-26. |
| [19] | 向小华, 吴新儒, 晁江涛, 等. 普通烟草WRKY基因家族的鉴定及表达分析[J]. 遗传, 2016, 38(9): 840-862. |
| Xiang XH, Wu XR, Chao JT, et al. Genome-wide identification and expression analysis of the WRKY gene family in common tobacco(Nicotiana tabacum L.)[J]. Hereditas, 2016, 38(9): 840-862. | |
| [20] | Lei RH, Li XL, Ma ZB, et al. Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function[J]. Plant J, 2017, 91(6): 962-976. |
| [21] | Zhao WH, Li YH, Fan SZ, et al. The transcription factor WRKY32 affects tomato fruit color by regulating YELLOW FRUITED-TOMATO 1, a core component of ethylene signal transduction[J]. J Exp Bot, 2021, 72(12): 4269-4282. |
| [22] | Zhang WW, Zhao SQ, Gu S, et al. FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in Fragaria vesca[J]. Plant Physiol, 2022, 189(2): 1037-1049. |
| [23] |
Li XY, Guo W, Li JC, et al. Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit[J]. Plant Physiol, 2020, 182(4): 2035-2046.
doi: 10.1104/pp.20.00002 pmid: 32047049 |
| [24] | Li A, Chen J, Lin Q, et al. Transcription factor MdWRKY32 participates in starch-sugar metabolism by binding to the MdBam5 promoter in apples during postharvest storage[J]. J Agric Food Chem, 2021, 69(49): 14906-14914. |
| [25] | 李欣容. 地黄WRKY家族基因鉴定及其在调控毛蕊花糖苷合成中的功能研究[D]. 郑州: 河南农业大学, 2021. |
| Li XR. Screening of WRKY family form Rehmannia glutinosa and their function in the biosynthesis of acteoside[D]. Zhengzhou: Henan Agricultural University, 2021. | |
| [26] | 张子轩. 萝卜黑色根皮花青素合成分子调控机制研究及相关基因克隆[D]. 太谷: 山西农业大学, 2022. |
| Zhang ZX. Molecular regulation mechanism and cloning related genes of anthocyanin synthesis in black root skin of radish[D]. Taigu: Shanxi Agricultural University, 2022. | |
| [27] |
赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0551 |
| Zhao ML, Guo YT, Ren YJ. Identification and analysis of WRKY transcription factor family genes in Helianthus tuberosus[J]. Biotechnol Bull, 2023, 39(2): 116-125. | |
| [28] | 赵令敏, 张艳芳, 邢丽南, 等. 山药异淀粉酶基因克隆及其在淀粉代谢中的作用[J]. 西北植物学报, 2022, 42(11): 1827-1834. |
| Zhao LM, Zhang YF, Xing LN, et al. Cloning of isoamylase gene in yam and its role in starch metabolism[J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(11): 1827-1834. | |
| [29] |
俞沁含, 李俊铎, 崔莹, 等. 山葡萄转录因子VaMYB4a互作蛋白的筛选与鉴定[J]. 园艺学报, 2023, 50(3): 508-522.
doi: 10.16420/j.issn.0513-353x.2021-1219 |
| Yu QH, Li JD, Cui Y, et al. Screening and identification of interacting protein of VaMYB4a from Vitis amurensis[J]. Acta Hortic Sin, 2023, 50(3): 508-522. | |
| [30] |
林胜男, 刘杰玮, 张晓妮, 等. 香石竹WRKY家族全基因组鉴定及其表达分析[J]. 园艺学报, 2021, 48(9): 1768-1784.
doi: 10.16420/j.issn.0513-353x.2019-0986 |
| Lin SN, Liu JW, Zhang XN, et al. Genome-wide identification and expression analysis of WRKY gene family in Dianthus caryophyllus[J]. Acta Hortic Sin, 2021, 48(9): 1768-1784. | |
| [31] | 任永娟, 王东姣, 苏亚春, 等. 植物WRKY转录因子: 结构、分类、进化和功能[J]. 农业生物技术学报, 2021, 29(1): 105-124. |
| Ren YJ, Wang DJ, Su YC, et al. Structure, classification, evolution and function of plant WRKY transcription factors[J]. J Agric Biotechnol, 2021, 29(1): 105-124. | |
| [32] | 蒋小刚. 甘蓝型油菜MADS-box基因家族鉴定及裂角调控网络初步分析[D]. 武汉: 华中农业大学, 2019. |
| Jiang XG. Identification of MADS-box gene family in Brassica napus L. and preliminary analysis of regulation network of pod dehiscence[D]. Wuhan: Huazhong Agricultural University, 2019. | |
| [33] | Liu J, Wang XY, Chen YL, et al. Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides[J]. Genomics, 2021, 113(5): 3294-3309. |
| [34] | 刘世芳, 刘霞宇, 张洁, 等. 甘薯IbWRKY75的克隆、亚细胞定位及表达特性分析[J]. 植物生理学报, 2020, 56(5): 969-980. |
| Liu SF, Liu XY, Zhang J, et al. Cloning, subcellular localization and expression analysis of IbWRKY75 in Ipomoea batatas[J]. Plant Physiol J, 2020, 56(5): 969-980. | |
| [35] | Tiika RJ, Wei J, Ma R, et al. Identification and expression analysis of the WRKY gene family during different developmental stages in Lycium ruthenicum Murr. fruit[J]. PeerJ, 2020, 8: e10207. |
| [36] | Xue CL, Li HT, Liu ZG, et al. Genome-wide analysis of the WRKY gene family and their positive responses to phytoplasma invasion in Chinese jujube[J]. BMC Genomics, 2019, 20(1): 464. |
| [37] | Chen CH, Chen XQ, Han J, et al. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses[J]. BMC Plant Biol, 2020, 20(1): 443. |
| [38] | Xu M, Wu C, Zhao LM, et al. WRKY transcription factor OpWRKY1 acts as a negative regulator of camptothecin biosynthesis in Ophiorrhiza pumila hairy roots[J]. Plant Cell Tissue Organ Cult, 2020, 142(1): 69-78. |
| [39] | 冯俊杰, 王远达, 邓琴霖, 等. 芥菜BjuWRKY75基因表达及其与开花整合子BjuFT互作[J]. 生物工程学报, 2022, 38(8): 3029-3040. |
| Feng JJ, Wang YD, Deng QL, et al. Expression of Brassica juncea BjuWRKY75 and its interactions with flowering integrator BjuFT[J]. Chin J Biotechnol, 2022, 38(8): 3029-3040. | |
| [40] | Zhang ZL, Xie Z, Zou XL, et al. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells[J]. Plant Physiol, 2004, 134(4): 1500-1513. |
| [41] | 张天亮. MdMYB 111和MdWRKY40参与红肉苹果花青苷生物合成的分子机制[D]. 泰安: 山东农业大学, 2020. |
| Zhang TL. Molecular mechanism of MdMYB111 and MdWRKY40 involved in anthocyanin biosynthesis in red- fleshed apple[D]. Tai'an: Shandong Agricultural University, 2020. | |
| [42] | Wang TJ, Huang SZ, Zhang A, et al. JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis[J]. New Phytol, 2021, 230(2): 567-584. |
| [43] | Ahmad R, Liu YT, Wang TJ, et al. GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid[J]. Plant Physiol, 2019, 179(4): 1844-1860. |
| [44] |
Van Aken O, Zhang BT, Law S, et al. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins[J]. Plant Physiol, 2013, 162(1): 254-271.
doi: 10.1104/pp.113.215996 pmid: 23509177 |
| [45] |
Wu SC, Blumer JM, Darvill AG, et al. Characterization of an endo-beta-1, 4-glucanase gene induced by auxin in elongating pea epicotyls[J]. Plant Physiol, 1996, 110(1): 163-170.
doi: 10.1104/pp.110.1.163 pmid: 8587980 |
| [46] |
Szyjanowicz PMJ, McKinnon I, Taylor NG, et al. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana[J]. Plant J, 2004, 37(5): 730-740.
doi: 10.1111/j.1365-313x.2003.02000.x pmid: 14871312 |
| [47] | Li H, Ye KY, Shi YT, et al. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis[J]. Mol Plant, 2017, 10(4): 545-559. |
| [48] |
Merlot S, Gosti F, Guerrier D, et al. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway[J]. Plant J, 2001, 25(3): 295-303.
pmid: 11208021 |
| [49] |
Doukhanina EV, Chen SR, van der Zalm E, et al. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana[J]. J Biol Chem, 2006, 281(27): 18793-18801.
doi: 10.1074/jbc.M511794200 pmid: 16636050 |
| [1] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [2] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [3] | WU Ze-hang, YANG Zhong-yi, YAN Yi-cheng, JIA Yong-hong, WU Yue-yan, XIE Xiao-hong. Cloning and Functional Analysis of Flavonoid 3'-hydroxylase(F3'H)Gene in Rhododendron hybridum Hort [J]. Biotechnology Bulletin, 2024, 40(6): 251-259. |
| [4] | WANG Qiu-yue, DUAN Peng-liang, LI Hai-xiao, LIU Ning, CAO Zhi-yan, DONG Jin-gao. Construction of cDNA Library of Setosphaeria turcica and Screening of Transcription Factor StMR1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(6): 281-289. |
| [5] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [6] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [7] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [8] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [9] | XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane [J]. Biotechnology Bulletin, 2024, 40(2): 197-211. |
| [10] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [11] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [12] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [13] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [14] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [15] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||