








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 106-117.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0247
Previous Articles Next Articles
NIE Zhu-xin1( ), GUO Jin1, QIAO Zi-yang1, LI Wei-wei1, ZHANG Xue-yan1, LIU Chun-yang1, WANG Jing1,2(
), GUO Jin1, QIAO Zi-yang1, LI Wei-wei1, ZHANG Xue-yan1, LIU Chun-yang1, WANG Jing1,2( )
)
Received:2024-03-14
Online:2024-08-26
Published:2024-07-31
Contact:
WANG Jing
E-mail:1538379214@qq.com;wangjing_imu@163.com
NIE Zhu-xin, GUO Jin, QIAO Zi-yang, LI Wei-wei, ZHANG Xue-yan, LIU Chun-yang, WANG Jing. Transcriptome Analysis of the Anthocyanin Biosynthesis in the Fruit Development Processes of Lycium ruthenicum Murr.[J]. Biotechnology Bulletin, 2024, 40(8): 106-117.
| 基因ID Gene ID | GenBank登录号GenBank number | 引物序列Primer sequence(5'-3') | 扩增长度Amplification length/bp |
|---|---|---|---|
| Cluster-53530 | — | F: ACCGATGAAGAACTCGTCGTCCA | 228 |
| R: GGTAGCCTTCCAATAACCCGATGT | |||
| Cluster-54040 | KY287796 | F: CTAAATGCCCCCAACCAGAACT | 124 |
| R: GTTACCCACTTCCCTTCATAGA | |||
| Cluster-55329 | OQ414189 | F: TTTTGGACTGCTGGTTCTTGTT | 260 |
| R: TTGAGGTCTTTGTTGATTATCGG | |||
| Cluster-57771 | — | F: CAAGGAGGAAATGCAAAGAGGAGC | 254 |
| R: CTATTAGGGACCATCTGTTGCCAAG | |||
| Cluster-59360 | — | F: CCGATGTAACGGATGGTGTCTA | 182 |
| R: GGGGCTGATTTGTCTCTATTGC | |||
| Cluster-67674 | KY287799 | F: AGAATGGGCACTGGCAGAAATGAT | 210 |
| R: ACTACACACGGCTCGTTTGATACC | |||
| Cluster-67748 | KF031378 | F: TCCCTTTTCCTTCCGAGTTCAT | 164 |
| R: TGTTTTGCCCTTCCACTGCTGC | |||
| Cluster-73437 | KY287797 | F: CACGGTAAGGAGACTGGTTTTCA | 167 |
| R: CAGCCTTCTCTGCCAGTATCTTG | |||
| Cluster-74326 | KY287798 | F: CAGTGGTGAACTCGGATAGCAGCA | 117 |
| R: GCTCCGCCATTACTGCTTTCTCTC | |||
| Cluster-76366 | — | F: GCAATCTCTTGTTCCTGGTTGTGAC | 288 |
| R: GGATACCCGCTGTTAGTGTTAGGAA | |||
| LrH2B1 | — | F: AGTGCTTCCTGGTGAATTGG | 163 |
| R: TGGATAATACCTAGCCCTAGTTTCC |
Table 1 Primer sequences used in this study
| 基因ID Gene ID | GenBank登录号GenBank number | 引物序列Primer sequence(5'-3') | 扩增长度Amplification length/bp |
|---|---|---|---|
| Cluster-53530 | — | F: ACCGATGAAGAACTCGTCGTCCA | 228 |
| R: GGTAGCCTTCCAATAACCCGATGT | |||
| Cluster-54040 | KY287796 | F: CTAAATGCCCCCAACCAGAACT | 124 |
| R: GTTACCCACTTCCCTTCATAGA | |||
| Cluster-55329 | OQ414189 | F: TTTTGGACTGCTGGTTCTTGTT | 260 |
| R: TTGAGGTCTTTGTTGATTATCGG | |||
| Cluster-57771 | — | F: CAAGGAGGAAATGCAAAGAGGAGC | 254 |
| R: CTATTAGGGACCATCTGTTGCCAAG | |||
| Cluster-59360 | — | F: CCGATGTAACGGATGGTGTCTA | 182 |
| R: GGGGCTGATTTGTCTCTATTGC | |||
| Cluster-67674 | KY287799 | F: AGAATGGGCACTGGCAGAAATGAT | 210 |
| R: ACTACACACGGCTCGTTTGATACC | |||
| Cluster-67748 | KF031378 | F: TCCCTTTTCCTTCCGAGTTCAT | 164 |
| R: TGTTTTGCCCTTCCACTGCTGC | |||
| Cluster-73437 | KY287797 | F: CACGGTAAGGAGACTGGTTTTCA | 167 |
| R: CAGCCTTCTCTGCCAGTATCTTG | |||
| Cluster-74326 | KY287798 | F: CAGTGGTGAACTCGGATAGCAGCA | 117 |
| R: GCTCCGCCATTACTGCTTTCTCTC | |||
| Cluster-76366 | — | F: GCAATCTCTTGTTCCTGGTTGTGAC | 288 |
| R: GGATACCCGCTGTTAGTGTTAGGAA | |||
| LrH2B1 | — | F: AGTGCTTCCTGGTGAATTGG | 163 |
| R: TGGATAATACCTAGCCCTAGTTTCC |
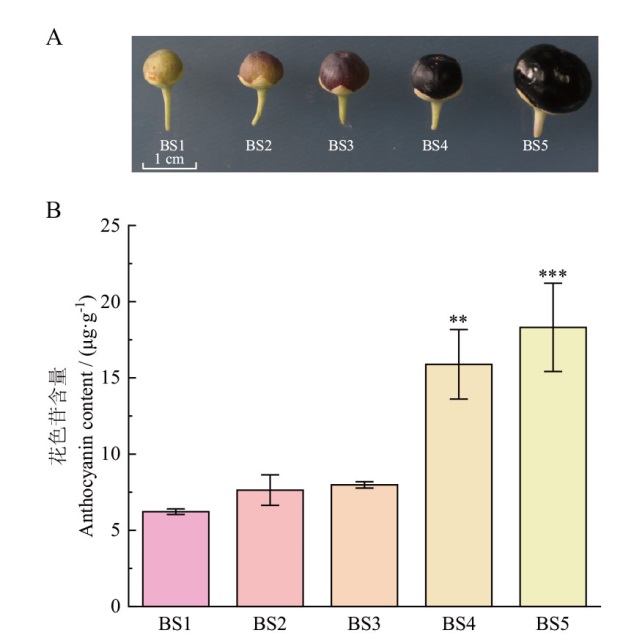
Fig. 1 Phenotypes(A)and anthocyanin contents(B)of fruit at different developmental stages of L. rutheni-cum Murr. BS1: Green fruit stage. BS2: Early color-changing stage. BS3: Late color-changing stage. BS4: Ripeness stage. BS5: Complete ripeness stage. Using BS1 as a control, ** P < 0.01, *** P < 0.001. The same below
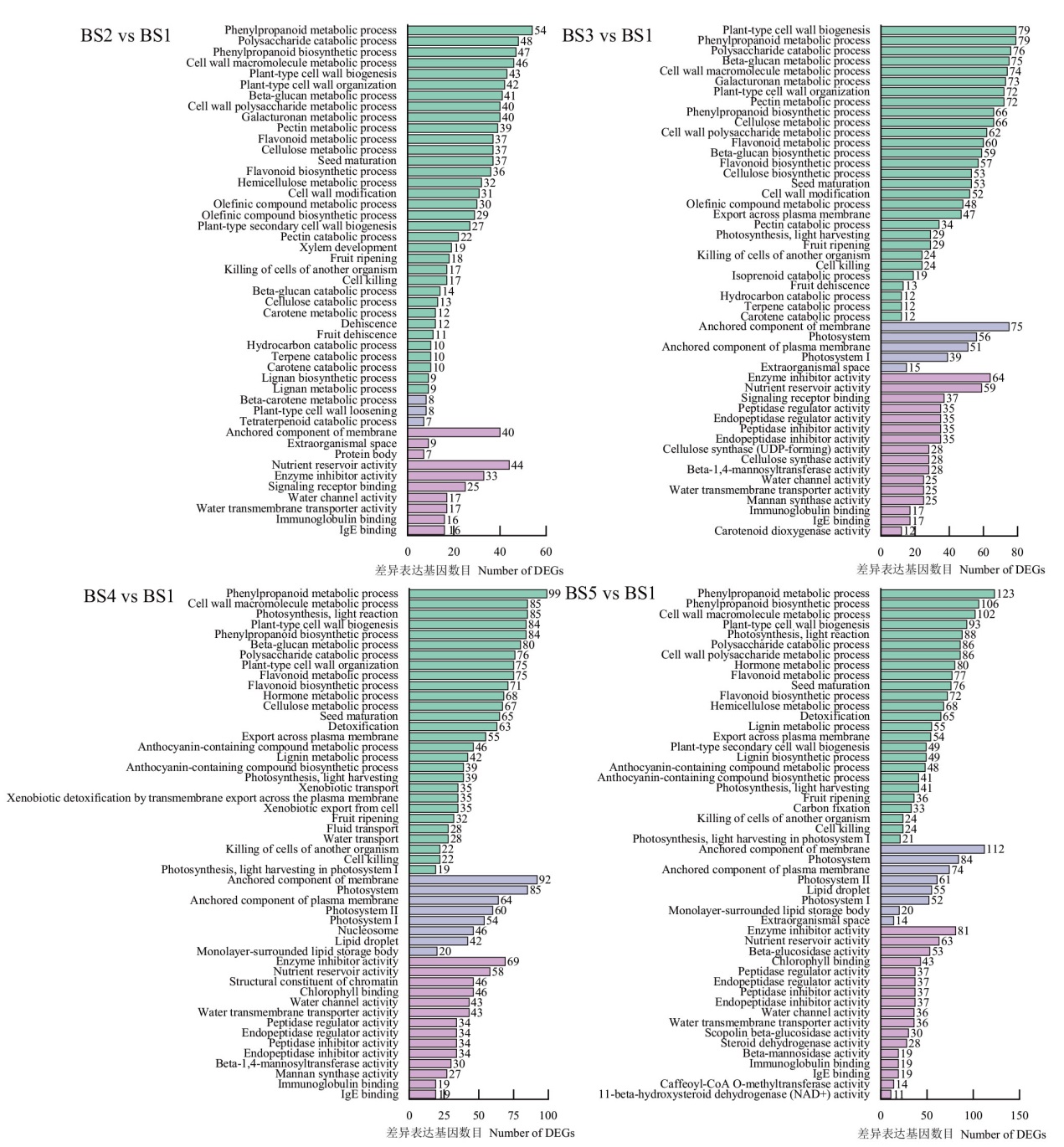
Fig. 3 Gene ontology annotation of DEGs in the fruits at different stages of L. ruthenicum Murr. Green: Biological process. Gray: Cell component. Purple: Molecular function
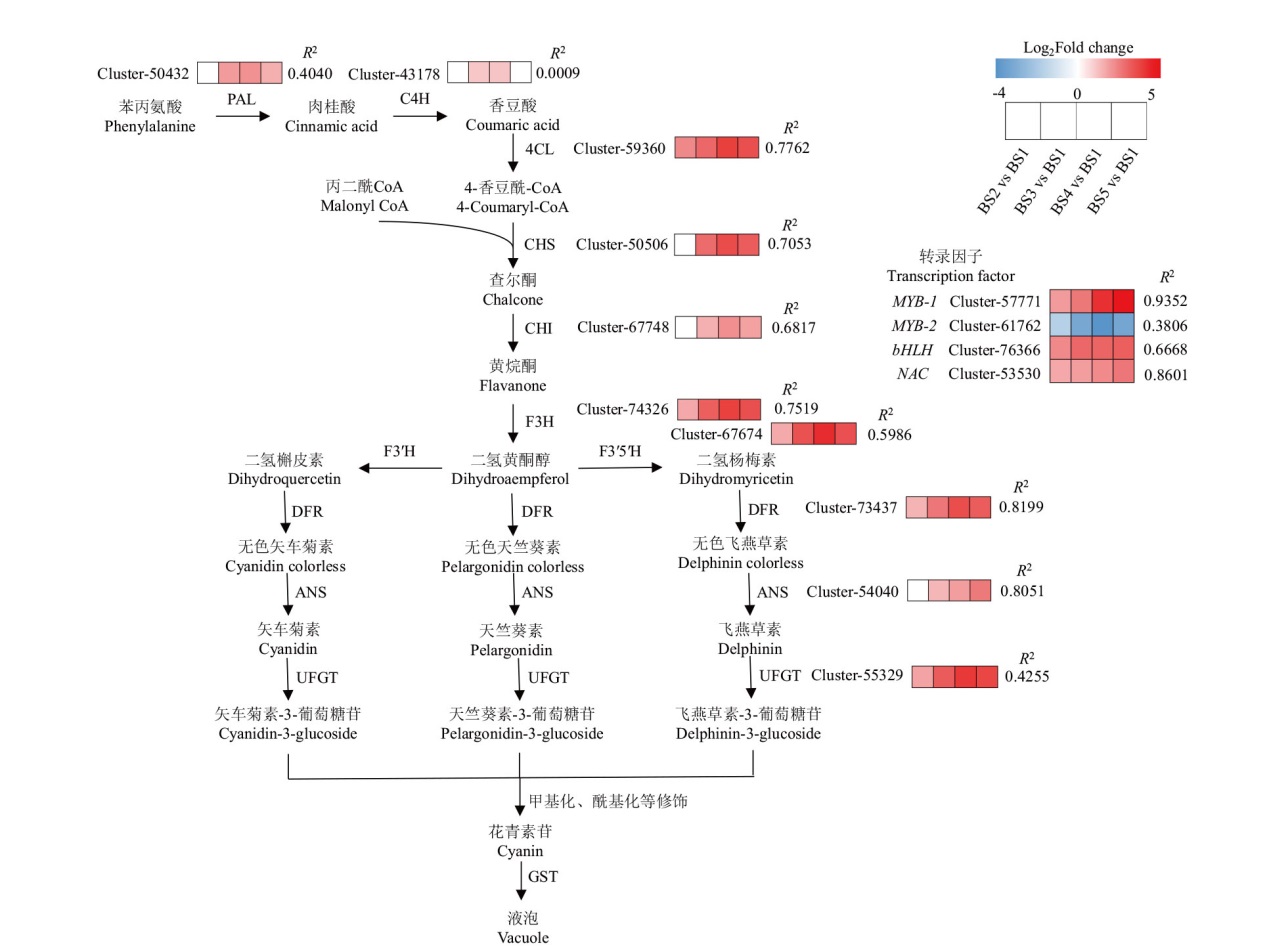
Fig. 6 Candidate genes related to anthocyanin biosynthesis in KEGG pathway and their correlation with anthocyanins R2: Correlation between gene expression(FPKM)and anthocyanin content. The same below
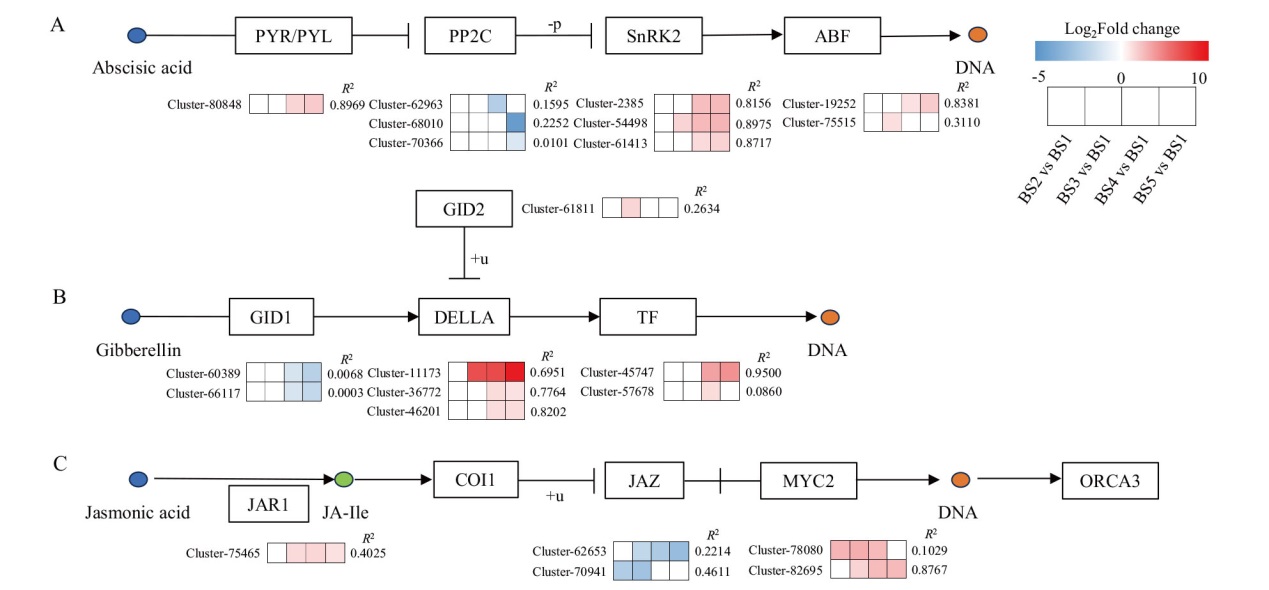
Fig. 7 Heat map of expressions of key genes in plant hormone signal transduction pathway A: ABA signal transduction pathway. B: GA signal transduction pathway. C: JA signal transduction pathway
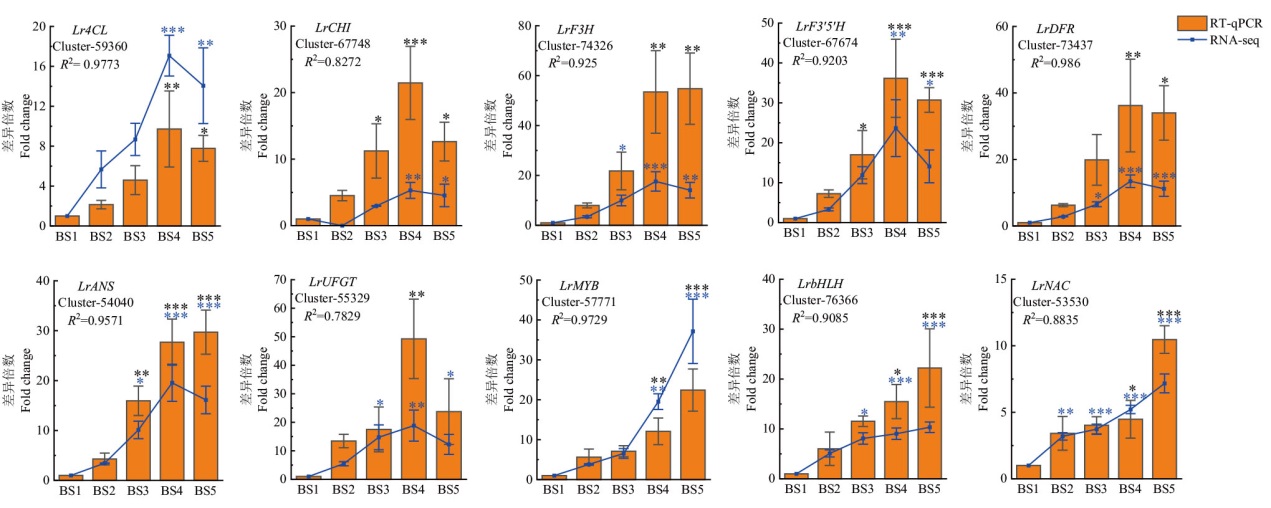
Fig. 8 Validation of transcriptome data Using BS1 as a control, * P < 0.05, ** P < 0.01, and *** P < 0.001. R2: Correlation between transcriptome data(RNA-seq)and RT-qPCR result(RT-qPCR)
| [1] | 薛晓敏. 套袋苹果果皮光照诱导后黑暗着色机理解析[D]. 杨凌: 西北农林科技大学, 2021. |
| Xue XM. Analysis of dark coloring mechanism of bagged apple peel induced by light[D]. Yangling: Northwest A & F University, 2021. | |
| [2] | 徐僡, 郑远静, 高方平, 等. 花色苷的生物合成及其影响因素研究进展[J]. 江苏农业学报, 2019, 35(5): 1246-1253. |
| Xu H, Zheng YJ, Gao FP, et al. Advances in the biosynthesis and influencing factors of anthocyanins[J]. Jiangsu J Agric Sci, 2019, 35(5): 1246-1253. | |
| [3] | Alappat B, Alappat J. Anthocyanin pigments: beyond aesthetics[J]. Molecules, 2020, 25(23): 5500. |
| [4] |
高国应, 伍小方, 张大为, 等. MBW复合体在植物花青素合成途径中的研究进展[J]. 生物技术通报, 2020, 36(1): 126-134.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0738 |
| Gao GY, Wu XF, Zhang DW, et al. Research progress on the MBW complexes in plant anthocyanin biosynthesis pathway[J]. Biotechnol Bull, 2020, 36(1): 126-134. | |
| [5] | 中国科学院中国植物志编辑委员会. 中国植物志[M]. 北京: 科学出版社, 1978. |
| Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora of China[M]. Beijing: Science Press, 1978. | |
| [6] | Wang HQ, Li JN, Tao WW, et al. Lycium ruthenicum studies: molecular biology, phytochemistry and pharmacology[J]. Food Chem, 2018, 240: 759-766. |
| [7] | Chen SS, Wang HL, Hu N. Long-term dietary Lycium ruthenicum Murr. anthocyanins intake alleviated oxidative stress-mediated aging-related liver injury and abnormal amino acid metabolism[J]. Foods, 2022, 11(21): 3377. |
| [8] | Zhang MJ, Xing LJ, Wang Y, et al. Anti-fatigue activities of anthocyanins from Lycium ruthenicum Murry[J]. Food Sci Technol, 2022, 42: e242703. |
| [9] |
林丽, 李进, 李永洁, 等. 黑果枸杞花色苷对氧化低密度脂蛋白损伤血管内皮细胞的保护作用[J]. 中国药学杂志, 2013, 48(8): 606-611.
doi: 10.11669/cpj.2013.08.008 |
| Lin L, Li J, Li YJ, et al. Protective effects of Lycium ruthenicum anthocyanins on the vascular endothelial cells with oxidative injury by oxidized low-density lipoprotein in vitro[J]. Chin Pharm J, 2013, 48(8): 606-611. | |
| [10] | Li ZL, Mi J, Lu L, et al. The main anthocyanin monomer of Lycium ruthenicum Murray induces apoptosis through the ROS/PTEN/PI3K/Akt/caspase3 signaling pathway in prostate cancer DU-145 cells[J]. Food Funct, 2021, 12(4): 1818-1828. |
| [11] | 武雪玲, 李筱筱, 贾世亮, 等. 黑果枸杞花青素对Aβ42致痴呆模型大鼠记忆力及抗氧化活性研究[J]. 现代食品科技, 2017, 33(3): 29-34. |
| Wu XL, Li XX, Jia SL, et al. Memory enhancing and antioxidant activities of Lycium ruthenicum Murray anthocyanin extracts in an Aβ42-induced rat model of dementia[J]. Mod Food Sci Technol, 2017, 33(3): 29-34. | |
| [12] |
孙燕, 王金刚, 臧丹丹, 等. 不同发育时期蓝果忍冬果实的转录组分析[J]. 生物技术通报, 2022, 38(12): 204-213.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0330 |
| Sun Y, Wang JG, Zang DD, et al. Transcriptome analysis of Lonicera caerulea fruits at different developmental stages[J]. Biotechnol Bull, 2022, 38(12): 204-213. | |
| [13] | Ma YJ, Duan HR, Zhang F, et al. Transcriptomic analysis of Lycium ruthenicum Murr. during fruit ripening provides insight into structural and regulatory genes in the anthocyanin biosynthetic pathway[J]. PLoS One, 2018, 13(12): e0208627. |
| [14] | Zong Y, Zhu XB, Liu ZG, et al. Functional MYB transcription factor encoding gene AN2 is associated with anthocyanin biosynthesis in Lycium ruthenicum Murray[J]. BMC Plant Biol, 2019, 19(1): 169. |
| [15] | Bao XM, Gan XL, Fan GH, et al. Transcriptome analysis identifies key genes involved in anthocyanin biosynthesis in black and purple fruits(Lycium ruthenicum Murr. L)[J]. Biotechnol Biotechnol Equip, 2022, 36(1): 553-560. |
| [16] |
郭嫒, 姜牧炎, 哈力马提·巴合太力, 等. 黑果枸杞茎叶响应NaCl胁迫合成花色苷的转录组学分析[J]. 生物技术通报, 2022, 38(10): 173-183.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1576 |
| Guo A, Jiang MY, HaLi-mati B, et al. Transcriptome analysis of Lycium ruthenicum Murr. shoots in anthocyanin biosynthesis response to salt stress[J]. Biotechnol Bull, 2022, 38(10): 173-183. | |
| [17] | Wang J, Jiang MY, Nie ZX, et al. ABA participates in salt stress-induced anthocyanin accumulation by stimulating the expression of LrMYB1 in Lycium ruthenicum Murr.[J]. Plant Cell Tissue Organ Cult, 2022, 151(1): 11-21. |
| [18] | Li G, Zhao JH, Qin BB, et al. ABA mediates development-dependent anthocyanin biosynthesis and fruit coloration in Lycium plants[J]. BMC Plant Biol, 2019, 19(1): 317. |
| [19] | Zeng SH, Wu M, Zou CY, et al. Comparative analysis of anthocyanin biosynthesis during fruit development in two Lycium species[J]. Physiol Plant, 2014, 150(4): 505-516. |
| [20] |
Lotkowska ME, Tohge T, Fernie AR, et al. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress[J]. Plant Physiol, 2015, 169(3): 1862-1880.
doi: 10.1104/pp.15.00605 pmid: 26378103 |
| [21] | Zhang SY, Ren YS, Zhao QY, et al. Drought-induced CsMYB6 interacts with CsbHLH111 to regulate anthocyanin biosynthesis in Chaenomeles speciosa[J]. Physiol Plant, 2023, 175(1): e13859. |
| [22] | Sun QG, Jiang SH, Zhang TL, et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11[J]. Plant Sci, 2019, 289: 110286. |
| [23] | 王会良, 何华平, 龚林忠, 等. 果树中花青苷合成研究进展[J]. 湖北农业科学, 2013, 52(20): 4857-4861. |
| Wang HL, He HP, Gong LZ, et al. Advances on the research of anthocyanin synthesis in fruit trees[J]. Hubei Agric Sci, 2013, 52(20): 4857-4861. | |
| [24] | 孟小伟, 牛赟, 马彦军. 黑果枸杞果实发育过程中转录组测序分析[J]. 中南林业科技大学学报, 2020, 40(9): 147-155. |
| Meng XW, Niu Y, Ma YJ. Transcriptome sequencing analysis of development of Lycium ruthenicum fruits[J]. J Cent South Univ For Technol, 2020, 40(9): 147-155. | |
| [25] | 张明菊, 朱莉, 夏启中. 植物激素对胁迫反应调控的研究进展[J]. 湖北大学学报: 自然科学版, 2021, 43(3): 242-253, 263. |
| Zhang MJ, Zhu L, Xia QZ. Research progress on the regulation of plant hormones to stress responses[J]. J Hubei Univ Nat Sci, 2021, 43(3): 242-253, 263. | |
| [26] | Han TY, Wu WL, Li WL. Transcriptome analysis revealed the mechanism by which exogenous ABA increases anthocyanins in blueberry fruit during veraison[J]. Front Plant Sci, 2021, 12: 758215. |
| [27] | Qu SS, Wang G, Li MM, et al. LcNAC90 transcription factor regulates biosynthesis of anthocyanin in harvested litchi in response to ABA and GA3[J]. Postharvest Biol Technol, 2022, 194: 112109. |
| [28] | Peng ZH, Han CY, Yuan LB, et al. Brassinosteroid enhances jasmonate-induced anthocyanin accumulation in Arabidopsis seedlings[J]. J Integr Plant Biol, 2011, 53(8): 632-640. |
| [29] | Shen SL, Hu XL, Cheng J, et al. PsbZIP1 and PsbZIP10 induce anthocyanin synthesis in plums(Prunus salicina cv. Taoxingli)via PsUFGT by methyl salicylate treatment during postharvest[J]. Postharvest Biol Technol, 2023, 203: 112396. |
| [30] | Kou YD, Ren JP, Ma YJ, et al. Effects of exogenous substances treatment on fruit quality and pericarp anthocyanin metabolism of peach[J]. Agronomy, 2023, 13(6): 1489. |
| [31] | An JP, Zhang XW, Liu YJ, et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple[J]. J Exp Bot, 2021, 72(4): 1460-1472. |
| [32] |
Jia HF, Jiu ST, Zhang C, et al. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor[J]. Plant Biotechnol J, 2016, 14(10): 2045-2065.
doi: 10.1111/pbi.12563 pmid: 27005823 |
| [33] | Samkumar A, Jones D, Karppinen K, et al. Red and blue light treatments of ripening bilberry fruits reveal differences in signalling through abscisic acid-regulated anthocyanin biosynthesis[J]. Plant Cell Environ, 2021, 44(10): 3227-3245. |
| [34] | Meng R, Zhang J, An L, et al. Expression profiling of several gene families involved in anthocyanin biosynthesis in apple(Malus domestica Borkh.)skin during fruit development[J]. J Plant Growth Regul, 2016, 35(2): 449-464. |
| [35] |
Zhang YQ, Liu ZJ, Liu JP, et al. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation[J]. Plant Cell Rep, 2017, 36(4): 557-569.
doi: 10.1007/s00299-017-2102-7 pmid: 28275852 |
| [36] | An JP, Xu RR, Liu X, et al. Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex[J]. Plant J, 2021, 106(5): 1414-1430. |
| [1] | ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata [J]. Biotechnology Bulletin, 2024, 40(8): 142-151. |
| [2] | LIAO Yang-mei, ZHAO Guo-chun, WENG Xue-huang, JIA Li-ming, CHEN Zhong. Transcriptome Sequencing of Male Sterile Buds at Different Developmental Stages in Sapindus mukorossi ‘Qirui’ [J]. Biotechnology Bulletin, 2024, 40(7): 197-206. |
| [3] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| [4] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [5] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [6] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [7] | WU Di, YOU Xiao-feng, ZHENG Yi-zheng, LIN Nan, ZHANG Yan-yan, WEI Yi-cong. Analysis of Endogenous Hormone Regulation Mechanism for Carotenoid Synthesis in Sarcandra glabra [J]. Biotechnology Bulletin, 2024, 40(5): 203-214. |
| [8] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [9] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [10] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [11] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [12] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| [13] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [14] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [15] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||