








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 129-141.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0179
Previous Articles Next Articles
YANG Wei1,2( ), ZHAO Li-fen1,2,3, TANG Bing1,2, ZHOU Lin-bi1,2, YANG Juan4, MO Chuan-yuan1,2, ZHANG Bao-hui1,2, LI Fei1,2, RUAN Song-lin5, DENG Ying1,2(
), ZHAO Li-fen1,2,3, TANG Bing1,2, ZHOU Lin-bi1,2, YANG Juan4, MO Chuan-yuan1,2, ZHANG Bao-hui1,2, LI Fei1,2, RUAN Song-lin5, DENG Ying1,2( )
)
Received:2024-02-24
Online:2024-08-26
Published:2024-07-31
Contact:
DENG Ying
E-mail:yangwei139@sina.cn;87928883@qq.com
YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L.[J]. Biotechnology Bulletin, 2024, 40(8): 129-141.
| 基因名称Gene name | 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| BjuVA1a | BjuVA05G09830.1 | CACTAAGTCACGCCGAGGAG | CTCTCCACCCACACCAAGTC |
| BjuVA1e | BjuVA09G34200.1 | AAACATCGAGGGGACGCAAA | AATGCTCCCCCAACTCCAAG |
| BjuVA2a | BjuVB03G32070.1 | ATGTAGGCCGCTTTTCCAGT | AACGTTTGCATCCCCTCGAT |
| BjuVA3a | BjuVA08G09520.1 | ACCCAGGATCCTCAAGGTGT | CTCATCTTGCAGCTTTCCGC |
| BjuVA4a | BjuVB07G38920.1 | GCGCTTATGAGACTCGCTCT | TCGCCCTCCTCGTAGATCAT |
| BjuVB1a | BjuVB03G28110.1 | TCACAACGAGAGCCAAGCAT | AGCCAACGTACCAAGCGTAT |
| BjuVB1b | BjuVA07G14180.1 | TCGATCCACTCTCCTCCTCC | GGAGAGGAAACACGTGGTGA |
| BjuVB2a | BjuVB04G02010.1 | CGCCGTGGAGAAAACAGAGT | AGAGCATTATCCGGGGAGAGA |
| ACT7 | NM_121018.4 | GGAATCGCTGACCGTATGAG | ACCCTCCAATCCAGACACTG |
Table 1 Primer sequences used in quantitative real-time PCR analysis of BjuSROs
| 基因名称Gene name | 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| BjuVA1a | BjuVA05G09830.1 | CACTAAGTCACGCCGAGGAG | CTCTCCACCCACACCAAGTC |
| BjuVA1e | BjuVA09G34200.1 | AAACATCGAGGGGACGCAAA | AATGCTCCCCCAACTCCAAG |
| BjuVA2a | BjuVB03G32070.1 | ATGTAGGCCGCTTTTCCAGT | AACGTTTGCATCCCCTCGAT |
| BjuVA3a | BjuVA08G09520.1 | ACCCAGGATCCTCAAGGTGT | CTCATCTTGCAGCTTTCCGC |
| BjuVA4a | BjuVB07G38920.1 | GCGCTTATGAGACTCGCTCT | TCGCCCTCCTCGTAGATCAT |
| BjuVB1a | BjuVB03G28110.1 | TCACAACGAGAGCCAAGCAT | AGCCAACGTACCAAGCGTAT |
| BjuVB1b | BjuVA07G14180.1 | TCGATCCACTCTCCTCCTCC | GGAGAGGAAACACGTGGTGA |
| BjuVB2a | BjuVB04G02010.1 | CGCCGTGGAGAAAACAGAGT | AGAGCATTATCCGGGGAGAGA |
| ACT7 | NM_121018.4 | GGAATCGCTGACCGTATGAG | ACCCTCCAATCCAGACACTG |
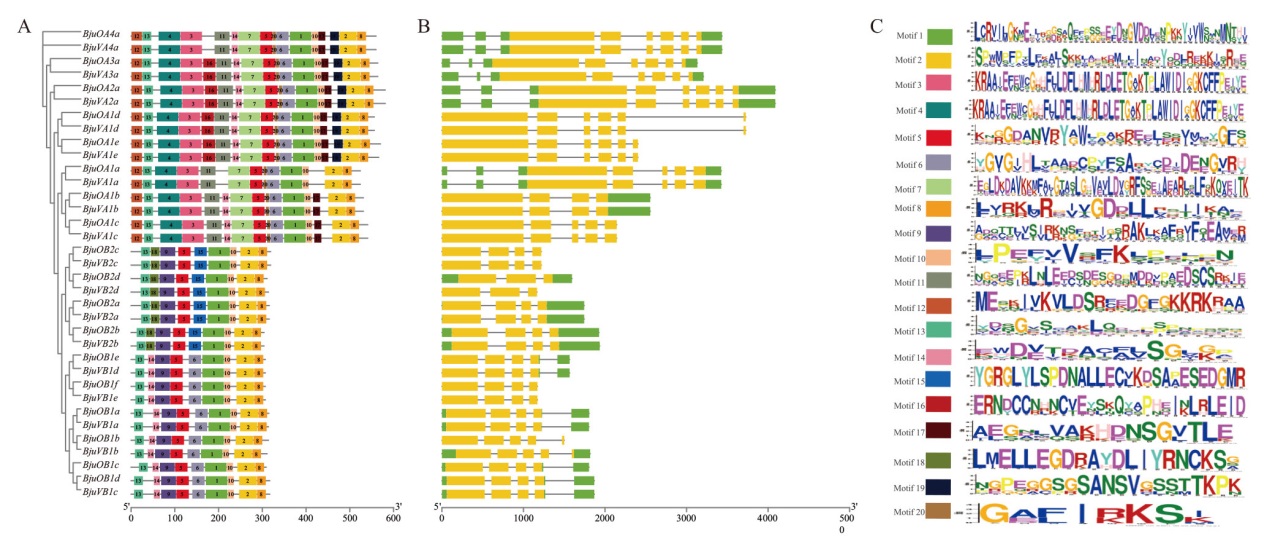
Fig. 1 Structure analysis of BjuSRO gene A: Conserved motif analysis of BjuSRO protein. B: Structure analysis of BjuSRO gene. C: Conserved motif logo of BjuSRO protein

Fig. 2 Domain analysis of BjuSRO protein A: Schematic diagram of SRO protein classification. B: 3D structure diagram of representative BjuSRO protein. C: PARP domain sequence alignment of BjuSRO protein. D: WWE domain sequence alignment of class A BjuSRO protein. E: SRT domain sequence alignment of BjuSRO protein
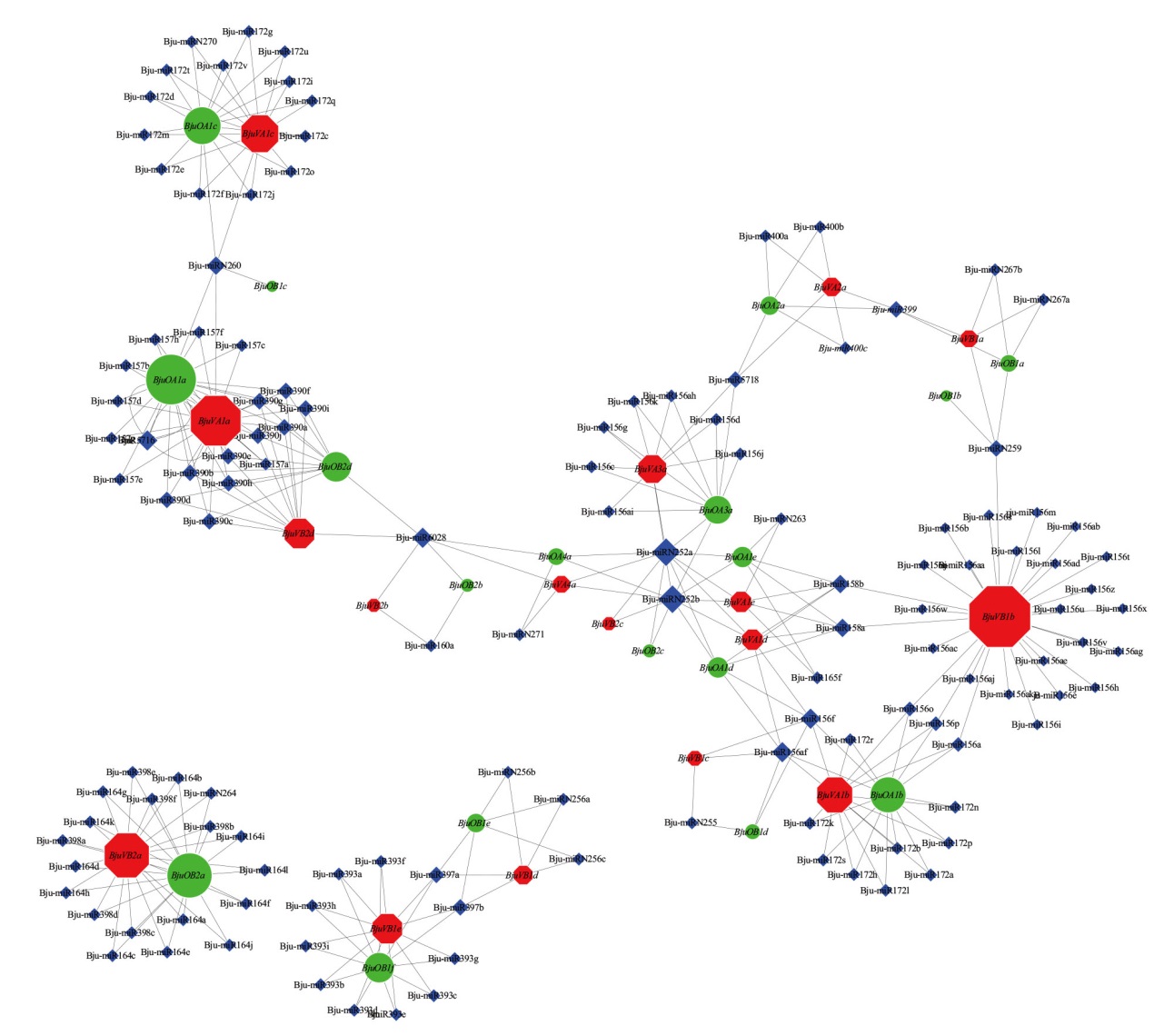
Fig. 4 Network diagram of interaction between Bju-miRNA and BjuSRO gene Diamonds indicate Bju-miRNAs; circles indicate BjuOSRO gene of oilseed mustard; octagon indicate BjuVSRO gene of vegetable mustard
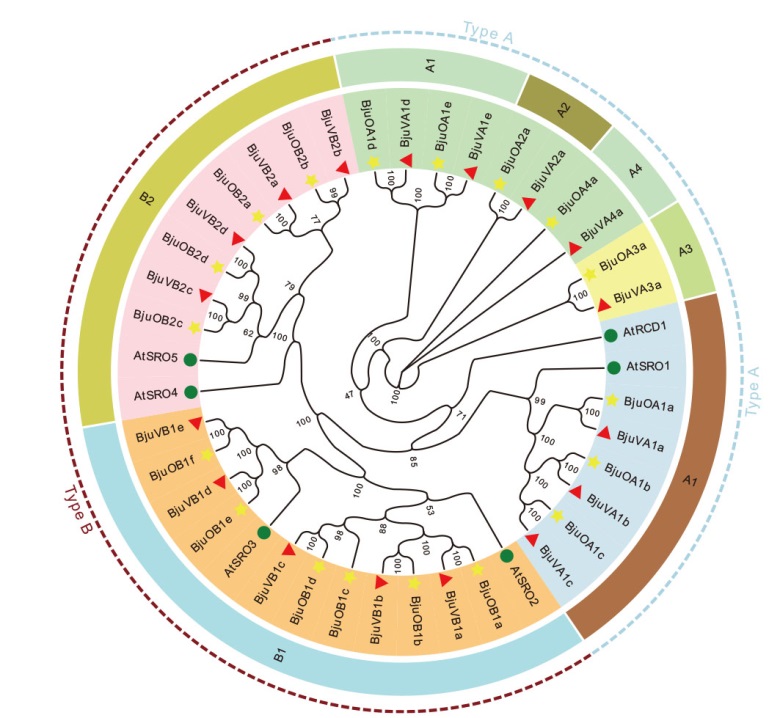
Fig. 7 Phylogenetic tree analysis of SRO gene family Circle indicates SRO proteins of Arabidopsis; pentagon indicate SRO proteins of oilseed mustard; triangle indicate SRO proteins of vegetable mustard
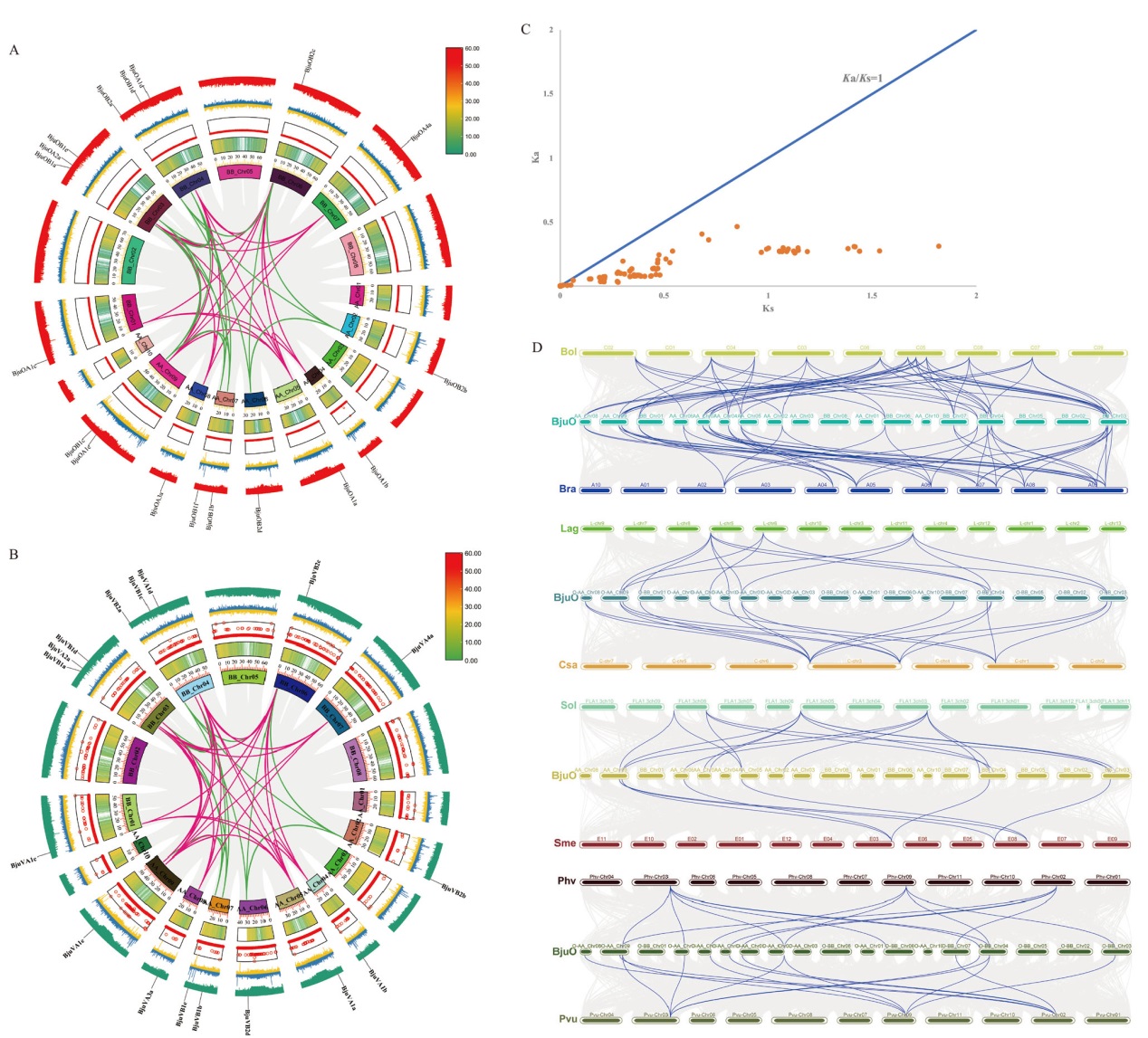
Fig. 8 Collinearity analysis of SRO gene A: Collinear analysis of BjuOSRO gene in oil mustard. B: Collinear analysis of BjuVSRO gene in vegetable mustard. Red line marks class A SRO gene, and green line marks class B SRO gene in Fig. A and B. C: Ka/Ks analysis of SRO homologous genes in mustard. D: Collinearity analysis of SRO gene between oil mustard and multi species(Bol: Cabbage; Bra: Chinese cabbage; Lag: loofah; Csa: cucumber; Sol: tomato; Sme: eggplant; Pvu: kidney beans; Phv: cowpea)
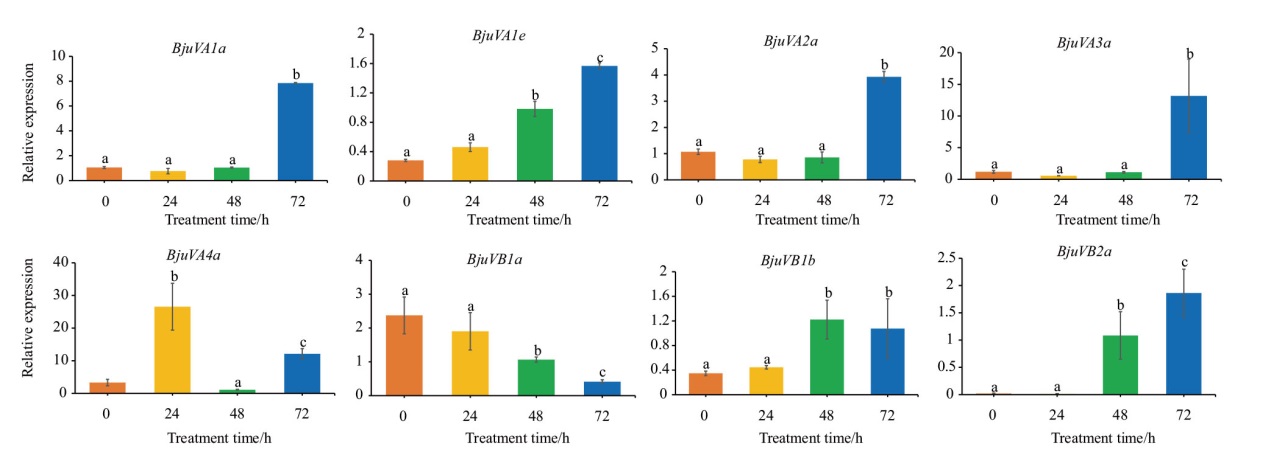
Fig. 9 Relative expressions of BjuSRO gene in mustard root system under salt stress at different times Different lowercase letters indicate significant differences(P<0.05)
| [1] | Zhang HM, Zhu JH, Gong ZZ, et al. Abiotic stress responses in plants[J]. Nat Rev Genet, 2022, 23(2): 104-119. |
| [2] |
Yang J, Ji LX, Liu S, et al. The CaM1-associated CCaMK-MKK1/6 cascade positively affects lateral root growth via auxin signaling under salt stress in rice[J]. J Exp Bot, 2021, 72(18): 6611-6627.
doi: 10.1093/jxb/erab287 pmid: 34129028 |
| [3] | Waadt R, Seller CA, Hsu PK, et al. Plant hormone regulation of abiotic stress responses[J]. Nat Rev Mol Cell Biol, 2022, 23(10): 680-694. |
| [4] | Chen H, Bullock DA Jr, Alonso JM, et al. To fight or to grow: the balancing role of ethylene in plant abiotic stress responses[J]. Plants, 2021, 11(1): 33. |
| [5] |
Chini A, Monte I, Fernández-Barbero G, et al. A small molecule antagonizes jasmonic acid perception and auxin responses in vascular and nonvascular plants[J]. Plant Physiol, 2021, 187(3): 1399-1413.
doi: 10.1093/plphys/kiab369 pmid: 34618088 |
| [6] | Zhang LC, He GH, Li YP, et al. PIL transcription factors directly interact with SPLs and repress tillering/branching in plants[J]. New Phytol, 2022, 233(3): 1414-1425. |
| [7] | Cheng ZY, Luan YT, Meng JS, et al. WRKY transcription factor response to high-temperature stress[J]. Plants, 2021, 10(10): 2211. |
| [8] | Liu J, Meng QL, Xiang HT, et al. Genome-wide analysis of Dof transcription factors and their response to cold stress in rice(Oryza sativa L.)[J]. BMC Genomics, 2021, 22(1): 800. |
| [9] |
Zhang HC, Shao SP, Zeng Y, et al. Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock[J]. Nat Cell Biol, 2022, 24(3): 340-352.
doi: 10.1038/s41556-022-00846-7 pmid: 35256776 |
| [10] | Verma S, Negi NP, Pareek S, et al. Auxin response factors in plant adaptation to drought and salinity stress[J]. Physiol Plant, 2022, 174(3): e13714. |
| [11] |
Alshareef NO, Otterbach SL, Allu AD, et al. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis[J]. Sci Rep, 2022, 12(1): 11264.
doi: 10.1038/s41598-022-14429-x pmid: 35787631 |
| [12] |
Jaspers P, Overmyer K, Wrzaczek M, et al. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants[J]. BMC Genomics, 2010, 11: 170.
doi: 10.1186/1471-2164-11-170 pmid: 20226034 |
| [13] | Jaspers P, Blomster T, Brosché M, et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors[J]. Plant J, 2009, 60(2): 268-279. |
| [14] |
Webb C, Upadhyay A, Giuntini F, et al. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway[J]. Biochemistry, 2011, 50(16): 3300-3309.
doi: 10.1021/bi2001888 pmid: 21417403 |
| [15] | Carter AA, Ramsey KM, Hatem CL, et al. Structural features of the Notch ankyrin domain-Deltex WWE2 domain heterodimer determined by NMR spectroscopy and functional implications[J]. Structure, 2023, 31(5): 584-594.e5. |
| [16] | Kim H, Aliar K, Tharmapalan P, et al. Differential DNA damage repair and PARP inhibitor vulnerability of the mammary epithelial lineages[J]. Cell Rep, 2023, 42(11): 113382. |
| [17] | Padella A, Ghelli Luserna Di Rorà A, Marconi G, et al. Targeting PARP proteins in acute leukemia: DNA damage response inhibition and therapeutic strategies[J]. J Hematol Oncol, 2022, 15(1): 10. |
| [18] | Maru B, Messikommer A, Huang LH, et al. PARP-1 improves leukemia outcomes by inducing parthanatos during chemotherapy[J]. Cell Rep Med, 2023, 4(9): 101191. |
| [19] |
Kong L, Feng BM, Yan Y, et al. Noncanonical mono(ADP-ribosyl)ation of zinc finger SZF proteins counteracts ubiquitination for protein homeostasis in plant immunity[J]. Mol Cell, 2021, 81(22): 4591-4604.e8.
doi: 10.1016/j.molcel.2021.09.006 pmid: 34592134 |
| [20] |
Vainonen JP, Jaspers P, Wrzaczek M, et al. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana[J]. Biochem J, 2012, 442(3): 573-581.
doi: 10.1042/BJ20111739 pmid: 22150398 |
| [21] |
Ahlfors R, Lång S, Overmyer K, et al. Arabidopsis radical-induced cell death1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses[J]. Plant Cell, 2004, 16(7): 1925-1937.
doi: 10.1105/tpc.021832 pmid: 15208394 |
| [22] | Xu W, Zhu WW, Yang L, et al. SMALL REPRODUCTIVE ORGANS, a SUPERMAN-like transcription factor, regulates stamen and pistil growth in rice[J]. New Phytol, 2022, 233(4): 1701-1718. |
| [23] |
You J, Zong W, Du H, et al. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors[J]. Plant Mol Biol, 2014, 84(6): 693-705.
doi: 10.1007/s11103-013-0163-8 pmid: 24337801 |
| [24] | Li HH, Li R, Qu FJ, et al. Identification of the SRO gene family in apples(Malus × domestica)with a functional characterization of MdRCD1[J]. Tree Genet Genomes, 2017, 13(5): 94. |
| [25] | Yuan BQ, Chen MH, Li SS. Isolation and identification of Ipomoea cairica(L.)sweet gene IcSRO1 encoding a SIMILAR TO RCD-ONE protein, which improves salt and drought tolerance in transgenic Arabidopsis[J]. Int J Mol Sci, 2020, 21(3): 1017. |
| [26] | Du YJ, Roldan MVG, Haraghi A, et al. Spatially expressed WIP genes control Arabidopsis embryonic root development[J]. Nat Plants, 2022, 8(6): 635-645. |
| [27] | Li XY, Xu YJ, Liu F, et al. Maize similar to RCD1 gene induced by salt enhances Arabidopsis thaliana abiotic stress resistance[J]. Biochem Biophys Res Commun, 2018, 503(4): 2625-2632. |
| [28] | Sanlier N, Guler SM. The benefits of Brassica vegetables on human health[J]. Journal of Human Health Research, 2018, 1(1): 104. |
| [29] | Yang JH, Wang J, Li ZP, et al. Genomic signatures of vegetable and oilseed allopolyploid Brassica juncea and genetic loci controlling the accumulation of glucosinolates[J]. Plant Biotechnol J, 2021, 19(12): 2619-2628. |
| [30] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [31] | Islam S, Shah SH, Corpas FJ, et al. Plant growth regulators mediated mitigation of salt-induced toxicities in mustard(Brassica juncea L.)by modifying the inherent defense system[J]. Plant Physiol Biochem, 2023, 196: 1002-1018. |
| [32] | Dixit S, Jangid VK, Grover A. Evaluation of suitable reference genes in Brassica juncea and its wild relative Camelina sativa for RT-qPCR analysis under various stress conditions[J]. PLoS One, 2019, 14(9): e0222530. |
| [33] |
Dizengremel P, Le Thiec D, Bagard M, et al. Ozone risk assessment for plants: central role of metabolism-dependent changes in reducing power[J]. Environ Pollut, 2008, 156(1): 11-15.
doi: 10.1016/j.envpol.2007.12.024 pmid: 18243452 |
| [34] | Li BZ, Zhao X, Zhao XL, et al. Structure and function analysis of Arabidopsis thaliana SRO protein family[J]. Yi Chuan, 2013, 35(10): 1189-1197. |
| [35] | Qiao YL, Gao XQ, Liu ZC, et al. Genome-wide identification and analysis of SRO gene family in Chinese cabbage(Brassica rapa L.)[J]. Plants, 2020, 9(9): 1235. |
| [36] | Vainonen JP, Shapiguzov A, Krasensky-Wrzaczek J, et al. Arabidopsis poly(ADP-ribose)-binding protein RCD1 interacts with photoregulatory protein kinases in nuclear bodies[J]. bioRxiv, 2020. https://doi.org/10.1101/2020.07.02.184937. |
| [37] | Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements[J]. Plant Sci, 2014, 217/218: 109-119. |
| [38] |
Lu TX, Rothenberg ME. MicroRNA[J]. J Allergy Clin Immunol, 2018, 141(4): 1202-1207.
doi: S0091-6749(17)31593-2 pmid: 29074454 |
| [39] |
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods[J]. J Cell Physiol, 2019, 234(5): 5451-5465.
doi: 10.1002/jcp.27486 pmid: 30471116 |
| [40] | Li N, Xu RQ, Wang BK, et al. Genome-wide identification and evolutionary analysis of the SRO gene family in tomato[J]. Front Genet, 2021, 12: 753638. |
| [41] |
Zhang L, Zhou DB, Hu HG, et al. Genome-wide characterization of a SRO gene family involved in response to biotic and abiotic stresses in banana(Musa spp.)[J]. BMC Plant Biol, 2019, 19(1): 211.
doi: 10.1186/s12870-019-1807-x pmid: 31113386 |
| [42] | Jiang WQ, Geng YP, Liu YK, et al. Genome-wide identification and characterization of SRO gene family in wheat: molecular evolution and expression profiles during different stresses[J]. Plant Physiol Biochem, 2020, 154: 590-611. |
| [43] | Qin LM, Sun L, Wei L, et al. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress[J]. Plant J, 2021, 105(4): 1010-1025. |
| [44] | Liu AL, Wei MY, Zhou Y, et al. Comprehensive analysis of SRO gene family in Sesamum indicum(L.)reveals its association with abiotic stress responses[J]. Int J Mol Sci, 2021, 22(23): 13048. |
| [45] |
Song XW, Li Y, Cao XF, et al. MicroRNAs and their regulatory roles in plant-environment interactions[J]. Annu Rev Plant Biol, 2019, 70: 489-525.
doi: 10.1146/annurev-arplant-050718-100334 pmid: 30848930 |
| [46] | Singh I, Smita S, Mishra DC, et al. Abiotic stress responsive miRNA-target network and related markers(SNP, SSR)in Brassica juncea[J]. Front Plant Sci, 2017, 8: 1943. |
| [47] | Wang YT, Wang RQ, Yu Y, et al. Genome-wide analysis of SIMILAR TO RCD ONE(SRO)family revealed their roles in abiotic stress in poplar[J]. Int J Mol Sci, 2023, 24(4): 4146. |
| [1] | ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata [J]. Biotechnology Bulletin, 2024, 40(8): 142-151. |
| [2] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [3] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [4] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [5] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [6] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [7] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [8] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [9] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [10] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [11] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [12] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [13] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [14] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [15] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||