








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 190-197.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0321
Previous Articles Next Articles
CUI Hai-yang( ), TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun(
), TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun( )
)
Received:2024-03-29
Online:2024-09-26
Published:2024-10-12
Contact:
TANG Li-chun
E-mail:chy@taiyuanshengwu.com;tlc@taiyuanshengwu.com
CUI Hai-yang, TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun. Generation of Virus-free TRAC-knocked-in T Cells Using Cas9TX[J]. Biotechnology Bulletin, 2024, 40(9): 190-197.
| 名称Name | 序列Sequence(5'-3') | |
|---|---|---|
| PCR Primers[ | TRAC-F4 | ATCACGAGCAGCTGGTTTCT |
| TRAC-R3 | GCCACCTTCTCTTCATCTGC | |
| qPCR Primers[ | TRAC-Up-F | GCATTTCAGGTTTCCTTGAGTGGCAG |
| TRAC-Up-R | TGGCAAGTCACGGTCTCATGCTTTAT | |
| TRAC-Cross-F | CTTGTCCATCACTGGCATCTGGACTC | |
| TRAC-Cross-R | ATCGGTGTGAATAGGCAGACAGACTTGT | |
| gRNAs[ | TRAC-A | ACAGATATCCAGAACCCTG |
| TRAC-R | TGTACCAGCTGAGAGACTCT | |
| TRAC-S | ACAAAACTGTGCTAGACATG | |
| ssDNA Templates[ | ssODN-F/ssODN-3'P | AGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGTCGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGC |
| ssODN-R/ssODN-R-3'P | GCTCCAGGCCACAGCACTGTTGCTCTTGAAGTCCATAGACCTCATGTCGACTAGCACAGTTTTGTCTGTGATATACACATCAGAATCCTTACT | |
Table 1 Sequences information
| 名称Name | 序列Sequence(5'-3') | |
|---|---|---|
| PCR Primers[ | TRAC-F4 | ATCACGAGCAGCTGGTTTCT |
| TRAC-R3 | GCCACCTTCTCTTCATCTGC | |
| qPCR Primers[ | TRAC-Up-F | GCATTTCAGGTTTCCTTGAGTGGCAG |
| TRAC-Up-R | TGGCAAGTCACGGTCTCATGCTTTAT | |
| TRAC-Cross-F | CTTGTCCATCACTGGCATCTGGACTC | |
| TRAC-Cross-R | ATCGGTGTGAATAGGCAGACAGACTTGT | |
| gRNAs[ | TRAC-A | ACAGATATCCAGAACCCTG |
| TRAC-R | TGTACCAGCTGAGAGACTCT | |
| TRAC-S | ACAAAACTGTGCTAGACATG | |
| ssDNA Templates[ | ssODN-F/ssODN-3'P | AGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGTCGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGC |
| ssODN-R/ssODN-R-3'P | GCTCCAGGCCACAGCACTGTTGCTCTTGAAGTCCATAGACCTCATGTCGACTAGCACAGTTTTGTCTGTGATATACACATCAGAATCCTTACT | |
| 组分Components | 用量Volume/μL |
|---|---|
| Nuclease Free ddH2O | 19.8 |
| NEB Buffer 2.1 | 2.5 |
| TRAC DNA Template | 2 |
| RNP | 0.7 |
| Total | 25 |
Table 2 Enzymatic digestion reaction system of RNP in vitro
| 组分Components | 用量Volume/μL |
|---|---|
| Nuclease Free ddH2O | 19.8 |
| NEB Buffer 2.1 | 2.5 |
| TRAC DNA Template | 2 |
| RNP | 0.7 |
| Total | 25 |
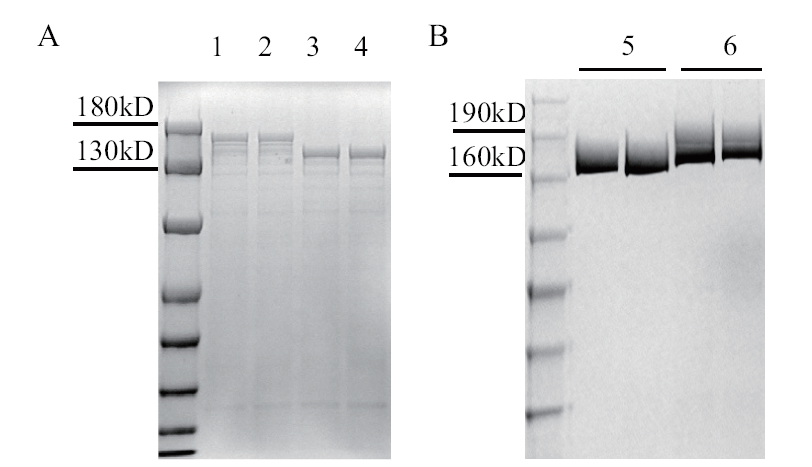
Fig. 1 Detection of Cas9 and Cas9TX protein by SDS-PAGE 1-2: Supernatant of Cas9TX bacterial lysate;3-4: supernatant of Cas9 bacterial lysate; 5: purified Cas9 protein; 6: purified Cas9TX protein
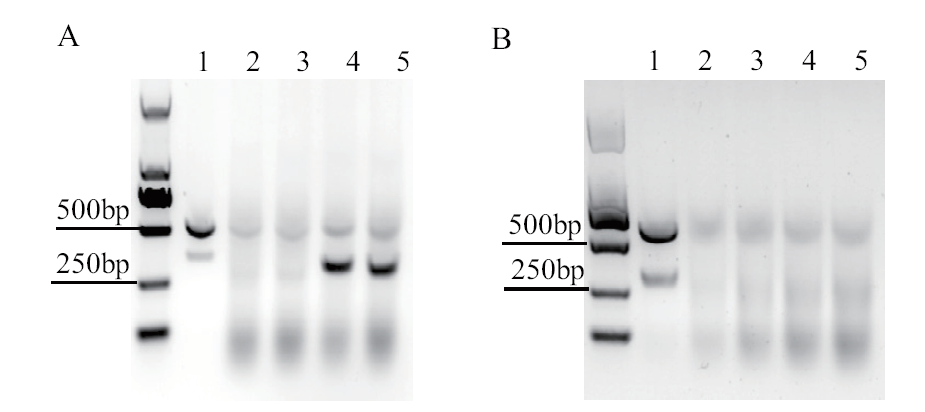
Fig. 2 Characterization of nuclease activity of Cas9 and Cas9TX in vitro A: RNP digestion of the TRAC DNA fragment. 1: target fragment; 2-3: target fragment with Cas9TX RNP; 4-5: target fragment with Cas9 RNP. B: Cut fragment with different concentrations of Cas9TX RNP. 1: 0.1 μmol/L; 2: 0.3 μmol/L; 3: 0.5 μmol/L; 4: 0.7 μmol/L; 5: 0.9 μmol/L
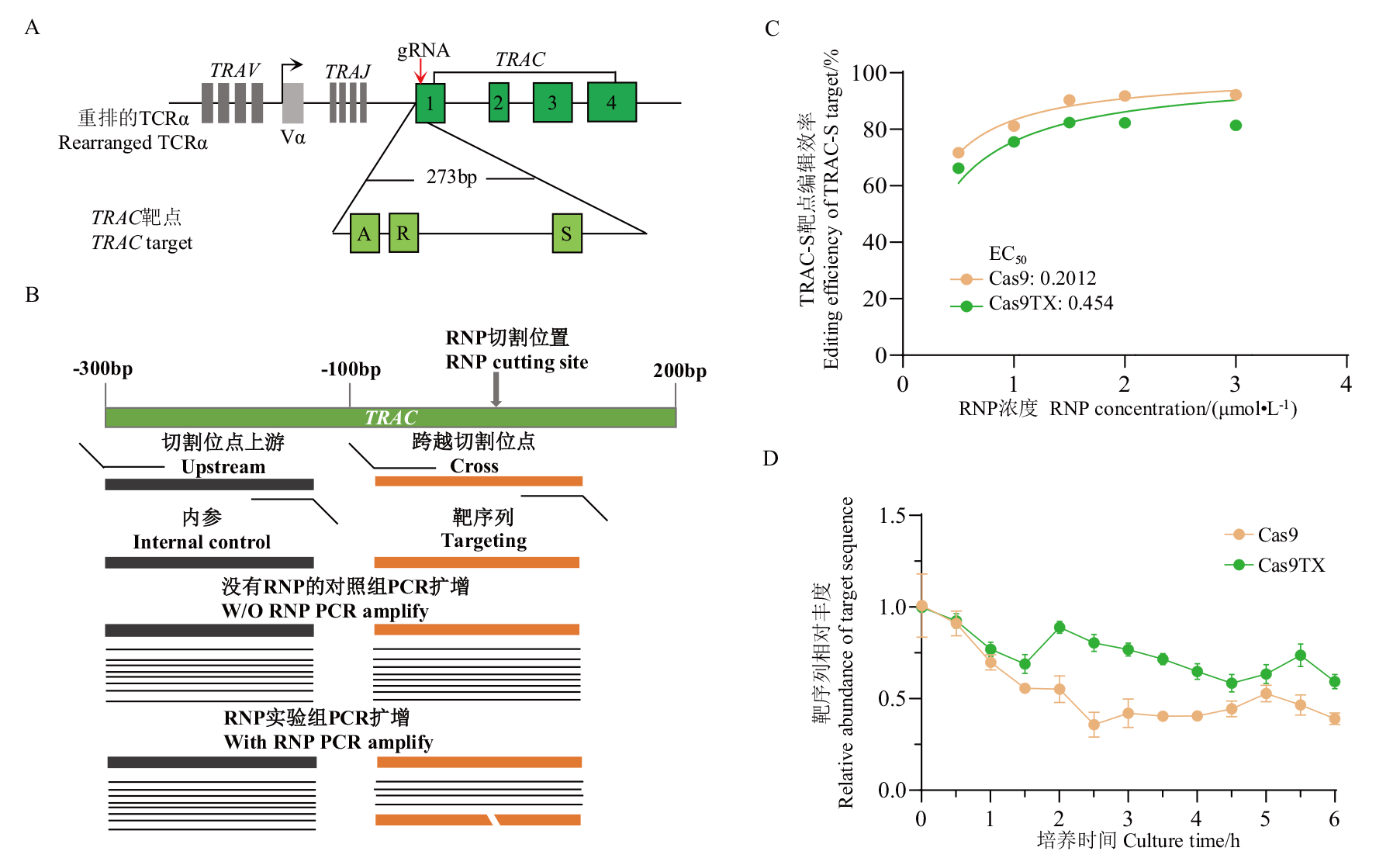
Fig. 3 Comparison of enzyme activity of Cas9 and Cas9TX in T cell A: gRNA targeted sites. B: Scheme of qPCR strategy to measure unlinked DSBs. C: EC50 curve. D: Kinetics curve of DSB generation

Fig. 4 Editing efficiencies of Cas9 and Cas9TX in different TRAC targets A: Knock out efficiency of Cas9 at TRAC-S site. B: Knock out efficiency of Cas9TX at TRAC-S site. C: Summary of knock out efficiencies at different targeted sites

Fig. 5 DNA donor templates can be degraded by Cas9TX and Cas9TX RNP A: dsDNA; B: ssODN; C: GW dsDNA; D: 3' P ssODN. 1, donor template; 2, donor template with Cas9; 3, donor template with Cas9TX; 4, donor template with Cas9 RNP; 5, donor template with Cas9TX RNP
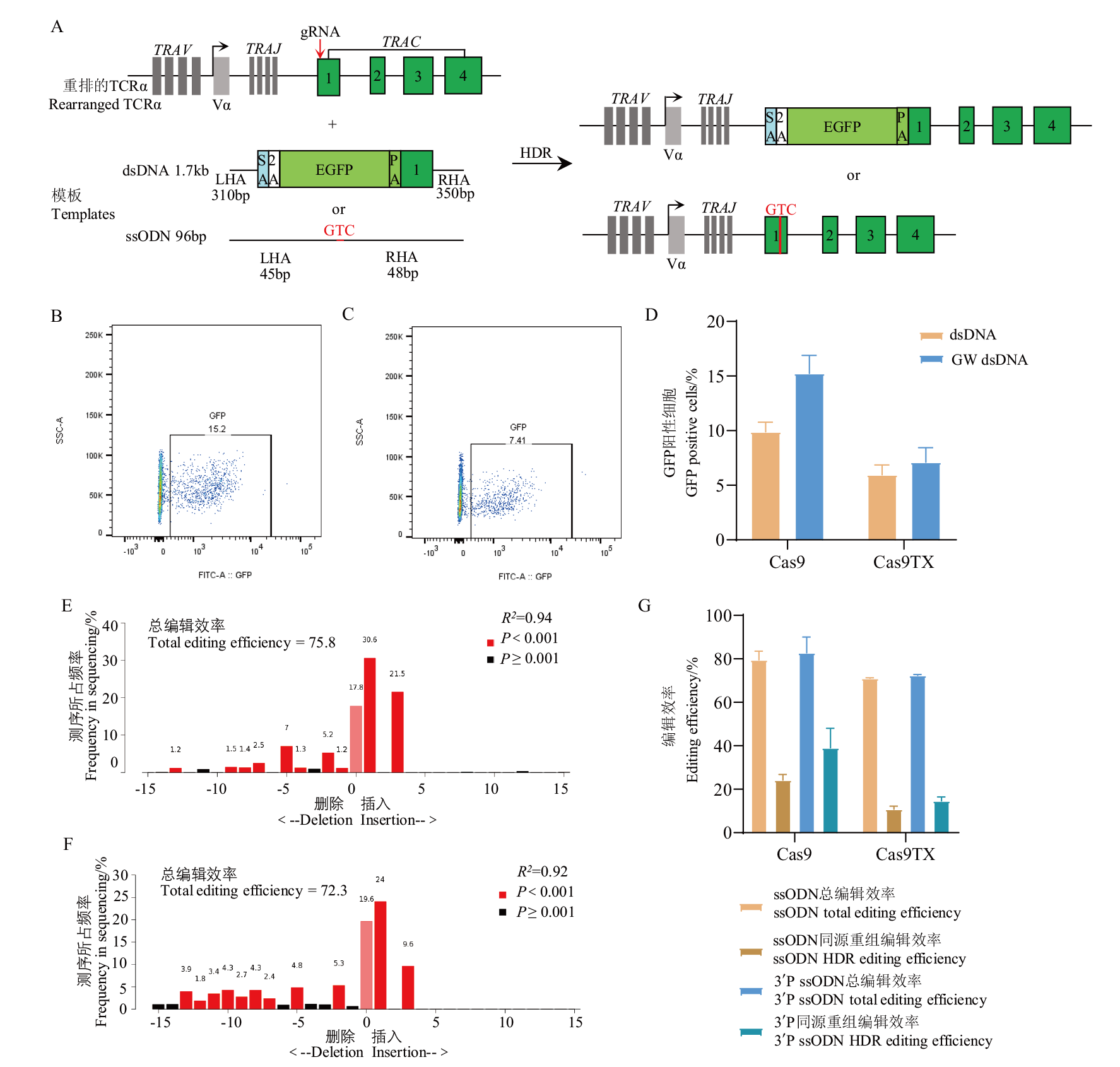
Fig. 6 Effects of donor-template modification on the site-directed insertion efficiencies A: Scheme of 2A-GFP or GTC knock in strategies; B: flow cytometric analysis for GFP expression using Cas9; C: flow cytometric analysis for GFP expression using Cas9TX; D: statistical results of GFP knock in efficiencies; E: TIDE analysis for GTC knock in efficiencies using Cas9; F: TIDE analysis for GTC knock in efficiencies using Cas9TX;G: statistical results of GTC knock in efficiencies
| [1] |
Larson RC, Maus MV. Recent advances and discoveries in the mechanisms and functions of CAR T cells[J]. Nat Rev Cancer, 2021, 21(3): 145-161.
doi: 10.1038/s41568-020-00323-z pmid: 33483715 |
| [2] |
Russo-Carbolante EM, Picanço-Castro V, Alves DCC, et al. Integration pattern of HIV-1 based lentiviral vector carrying recombinant coagulation factor VIII in Sk-Hep and 293T cells[J]. Biotechnol Lett, 2011, 33(1): 23-31.
doi: 10.1007/s10529-010-0387-5 pmid: 20812025 |
| [3] |
Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system[J]. J Immunol, 2013, 190(5): 1911-1918.
doi: 10.4049/jimmunol.1203162 pmid: 23417527 |
| [4] |
Monjezi R, Miskey C, Gogishvili T, et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors[J]. Leukemia, 2017, 31(1): 186-194.
doi: 10.1038/leu.2016.180 pmid: 27491640 |
| [5] |
Foster JB, Choudhari N, Perazzelli J, et al. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response[J]. Hum Gene Ther, 2019, 30(2): 168-178.
doi: 10.1089/hum.2018.145 pmid: 30024272 |
| [6] |
Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements[J]. Nat Biotechnol, 2018, 36(8): 765-771.
doi: 10.1038/nbt.4192 pmid: 30010673 |
| [7] | Casini A, Olivieri M, Petris G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast[J]. Nat Biotechnol, 2018, 36(3): 265-271. |
| [8] |
Chen BH, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system[J]. Cell, 2013, 155(7): 1479-1491.
doi: 10.1016/j.cell.2013.12.001 pmid: 24360272 |
| [9] |
Yu Y, Guo YJ, Tian QQ, et al. An efficient gene knock-in strategy using 5'-modified double-stranded DNA donors with short homology arms[J]. Nat Chem Biol, 2020, 16(4): 387-390.
doi: 10.1038/s41589-019-0432-1 pmid: 31873222 |
| [10] |
Yin JH, Lu RS, Xin CC, et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing[J]. Nat Commun, 2022, 13(1): 1204.
doi: 10.1038/s41467-022-28900-w pmid: 35260581 |
| [11] |
Yin JH, Fang KL, Gao YX, et al. Safeguarding genome integrity during gene-editing therapy in a mouse model of age-related macular degeneration[J]. Nat Commun, 2022, 13(1): 7867.
doi: 10.1038/s41467-022-35640-4 pmid: 36550137 |
| [12] | Kath J, Du WJ, Pruene A, et al. Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR Tcells[J]. Mol Ther Methods Clin Dev, 2022, 25: 311-330. |
| [13] |
Liu XJ, Zhang YP, Cheng C, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells[J]. Cell Res, 2017, 27(1): 154-157.
doi: 10.1038/cr.2016.142 pmid: 27910851 |
| [14] | Yang M, Tkach D, Boyne A, et al. Optimized two-step electroporation process to achieve efficient nonviral-mediated gene insertion into primary T cells[J]. FEBS Open Bio, 2022, 12(1): 38-50. |
| [15] |
Harrigan JA, Fan JS, Momand J, et al. WRN exonuclease activity is blocked by DNA termini harboring 3' obstructive groups[J]. Mech Ageing Dev, 2007, 128(3): 259-266.
pmid: 17224176 |
| [16] |
Liao Q, Zheng JW, Wang B, et al. Efficient soluble expression and application of SpCas9 protein[J]. Food Science, 2023, 44(10): 150-157.
doi: 10.7506/spkx1002-6630-20220429-391 |
| [17] | 刘芳, 卢婷, 蔡梦迪, 等. Cas9蛋白的克隆表达、分离纯化及多克隆抗体制备[J]. 贵州医科大学学报, 2019, 44(7): 757-761, 766. |
| Liu F, Lu T, Cai MD, et al. Cloning expression, purification and polyclonal antibody preparation of Cas9 protein[J]. J Guizhou Med Univ, 2019, 44(7): 757-761, 766. | |
| [18] | Hu JZ, Yin JX. Fusion protein and use method thereof: WO2023011638(A1)[P]. 2023-02-09. |
| [1] | HOU Wen-ting, SUN Lin, ZHANG Yan-jun, DONG He-zhong. Application of Gene-editing Technology for Germplasm Innovation and Genetic Improvement in Cotton [J]. Biotechnology Bulletin, 2024, 40(7): 68-77. |
| [2] | ZHU Tian-yi, KONG Gui-mei, JIAO Hong-mei, GUO Ting-ting, WU Ri-han, LIU Cui-cui, GAO Cheng-feng, LI Guo-cai. Establishment of A Bacterial Model of CRISPR/Cas9 Mediated adeG Gene Knockout in Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(2): 55-64. |
| [3] | GAO Deng-ke, MA Bai-rong, GUO Yi-ying, LIU Wei, LIU Tian, JIN Ya-ping, JIANG Zhou, CHEN Hua-tao. Establishment of Quaking Knockout Mouse Embryonic Fibroblast Cell Line Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2024, 40(2): 65-72. |
| [4] | ZHANG Hong-min, LONG Wen, LAO Xiao-qing, CHEN Wen-yan, SHANG Xue-mei, WANG Hong-lian, WANG Li, SU Hong-wei, SHEN Hong-ping, SHEN Hong-chun. Construction of Pmepa1 Knockout TCMK1 Mouse Renal Tubular Epithelial Cell Line Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2024, 40(2): 73-79. |
| [5] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [6] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [7] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [8] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [9] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [10] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [11] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [12] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [13] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [14] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [15] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||