








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 198-211.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0269
Previous Articles Next Articles
WU Hui-qin( ), WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan(
), WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan( )
)
Received:2024-03-19
Online:2024-09-26
Published:2024-10-12
Contact:
CHENG Yan
E-mail:1993148688@qq.com;chengyan820620@163.com
WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper[J]. Biotechnology Bulletin, 2024, 40(9): 198-211.
| 基因 Gene | 正向引物Forward primer(5'-3') | 正向引物Forward primer(5'-3') |
|---|---|---|
| CaUGT31 | GCTACAAAGCTGATGCAGGC | GCGAAGACACTCGTACCGAA |
| CaUGT39 | AAGGAGGCCATCGACACAAG | TAGGCAGGATCAAGCTGTGC |
| CaUGT47 | GGGCCTTCTTCCACGAATTT | CGATCCCAAGTCCTCCAAGTT |
| CaUGT53 | GGTTCTTGACTCACAGCGGA | GCGAAAAATGGCCAGCAGAT |
| CaUGT63 | TTAGCTTTGGGAGTGTCGCC | ATCCGCGGGAATCTTGGTTT |
| CaUGT74 | GCTCCTCTTCCCTCTTCCAA | CAAACCGGCGAGACAAAAGA |
| CaUGT82 | CTGAGATGGCATGGGGACTT | GGCAAATTTACAACGTCGTCC |
| CaUGT92 | TGCCGTGTCAATTCGAAAGG | GCAACTTCAACAGCCCATGA |
| CaUGT127 | ATGTCCAAACCCAACTTCACA | CACGGCAGCACCAAAAGTTA |
| CaUGT128 | CCAATGAATGCCAGGTTGGTAG | TGGCCCTCAAATTTTCCCCCAT |
| CaUGT132 | GCCCCTCAAGCAACAATTCT | CACTGGTTGGTCAATGTGCA |
| CaActin | GGTGACGAGGCTCAATCCAA | CTCTGGAGCCACACGAAGTT |
Table 1 RT-qPCR primer sequences of CaUGTs
| 基因 Gene | 正向引物Forward primer(5'-3') | 正向引物Forward primer(5'-3') |
|---|---|---|
| CaUGT31 | GCTACAAAGCTGATGCAGGC | GCGAAGACACTCGTACCGAA |
| CaUGT39 | AAGGAGGCCATCGACACAAG | TAGGCAGGATCAAGCTGTGC |
| CaUGT47 | GGGCCTTCTTCCACGAATTT | CGATCCCAAGTCCTCCAAGTT |
| CaUGT53 | GGTTCTTGACTCACAGCGGA | GCGAAAAATGGCCAGCAGAT |
| CaUGT63 | TTAGCTTTGGGAGTGTCGCC | ATCCGCGGGAATCTTGGTTT |
| CaUGT74 | GCTCCTCTTCCCTCTTCCAA | CAAACCGGCGAGACAAAAGA |
| CaUGT82 | CTGAGATGGCATGGGGACTT | GGCAAATTTACAACGTCGTCC |
| CaUGT92 | TGCCGTGTCAATTCGAAAGG | GCAACTTCAACAGCCCATGA |
| CaUGT127 | ATGTCCAAACCCAACTTCACA | CACGGCAGCACCAAAAGTTA |
| CaUGT128 | CCAATGAATGCCAGGTTGGTAG | TGGCCCTCAAATTTTCCCCCAT |
| CaUGT132 | GCCCCTCAAGCAACAATTCT | CACTGGTTGGTCAATGTGCA |
| CaActin | GGTGACGAGGCTCAATCCAA | CTCTGGAGCCACACGAAGTT |
| 项目 Item | 蛋白长度 Protein length/aa | 分子量 Molecular weight/kD | 等电点 pI | 不稳定指数 Instability index | 疏水指数 GRAVY | 带负电残基数 Asp + Glu | 带正电残基数 Arg + Lys |
|---|---|---|---|---|---|---|---|
| 分布范围 | 253-534 | 20.71×103-59.65×103 | 4.7-8.45 | 26.52-59.99 | -0.334-0.054 | 29-73 | 20-64 |
| 平均值 | 450 | 50.61×103 | 5.79 | 43.35 | -0.139 | 55.53 | 45.86 |
| 亲水蛋白数 | — | — | — | — | 131 | — | — |
| 疏水蛋白数 | — | — | — | — | 9 | — | — |
| 酸性蛋白数 | — | — | 134 | — | — | — | — |
| 碱性蛋白数 | — | — | 6 | — | — | — | — |
| 稳定蛋白 | — | — | — | 40 | — | — | — |
| 不稳定蛋白 | — | — | — | 100 | — | — | — |
| 最小值成员 | CaUGT43 | CaUGT43 | CaUGT101 | CaUGT102 | CaUGT67 | CaUGT15 | CaUGT137 |
| 最大值成员 | CaUGT99 | CaUGT99 | CaUGT137 | CaUGT61 | CaUGT49 | CaUGT91 | CaUGT49 |
Table 2 Physical and chemical properties of CaUGTs protein
| 项目 Item | 蛋白长度 Protein length/aa | 分子量 Molecular weight/kD | 等电点 pI | 不稳定指数 Instability index | 疏水指数 GRAVY | 带负电残基数 Asp + Glu | 带正电残基数 Arg + Lys |
|---|---|---|---|---|---|---|---|
| 分布范围 | 253-534 | 20.71×103-59.65×103 | 4.7-8.45 | 26.52-59.99 | -0.334-0.054 | 29-73 | 20-64 |
| 平均值 | 450 | 50.61×103 | 5.79 | 43.35 | -0.139 | 55.53 | 45.86 |
| 亲水蛋白数 | — | — | — | — | 131 | — | — |
| 疏水蛋白数 | — | — | — | — | 9 | — | — |
| 酸性蛋白数 | — | — | 134 | — | — | — | — |
| 碱性蛋白数 | — | — | 6 | — | — | — | — |
| 稳定蛋白 | — | — | — | 40 | — | — | — |
| 不稳定蛋白 | — | — | — | 100 | — | — | — |
| 最小值成员 | CaUGT43 | CaUGT43 | CaUGT101 | CaUGT102 | CaUGT67 | CaUGT15 | CaUGT137 |
| 最大值成员 | CaUGT99 | CaUGT99 | CaUGT137 | CaUGT61 | CaUGT49 | CaUGT91 | CaUGT49 |
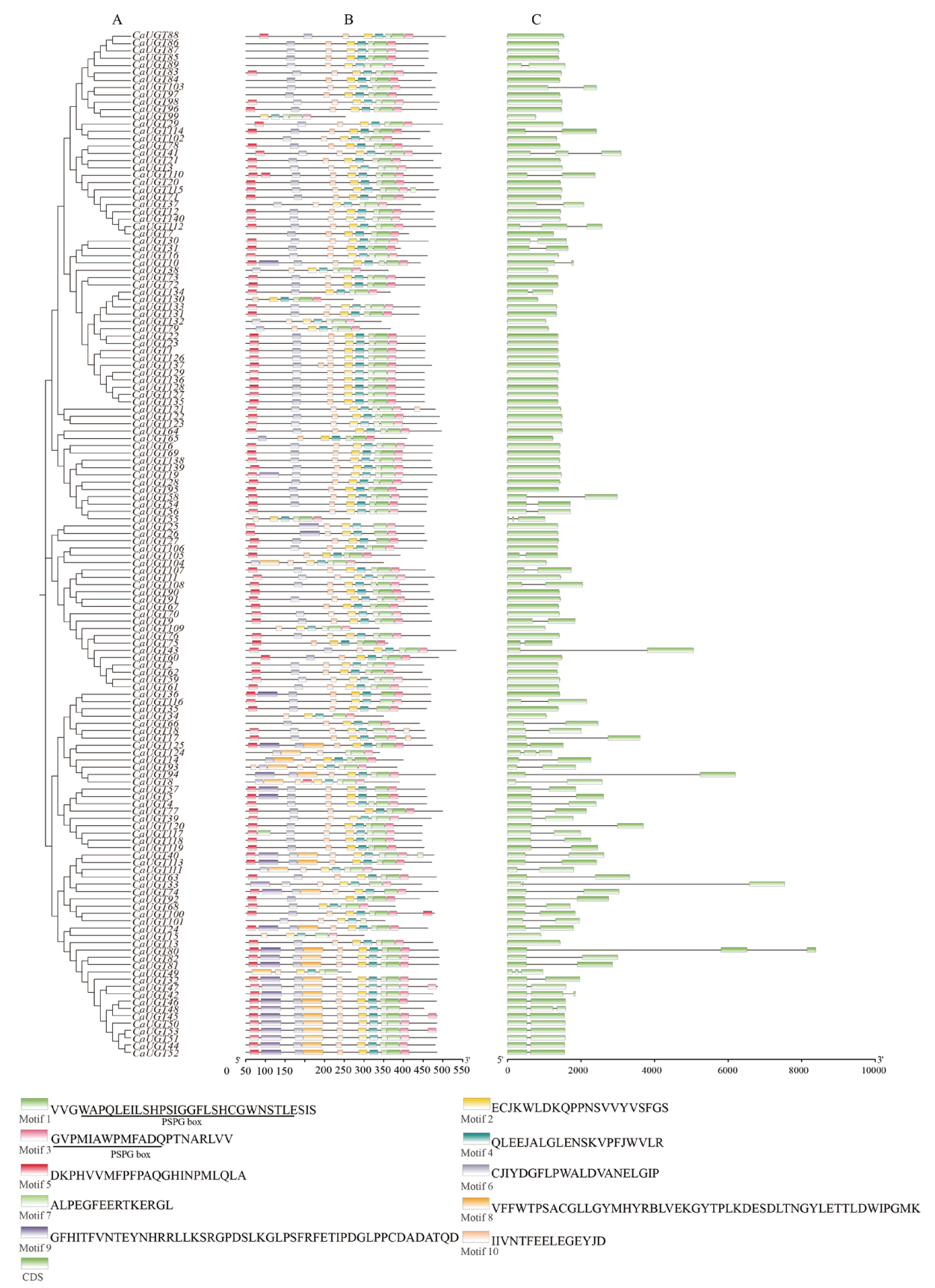
Fig. 2 Phylogngntic tree, gene structure and motif distribution of the pepper CaUGTs family member A: Phylogenetic tree; B: conserved domain composition; C: gene structure(exon/intron)
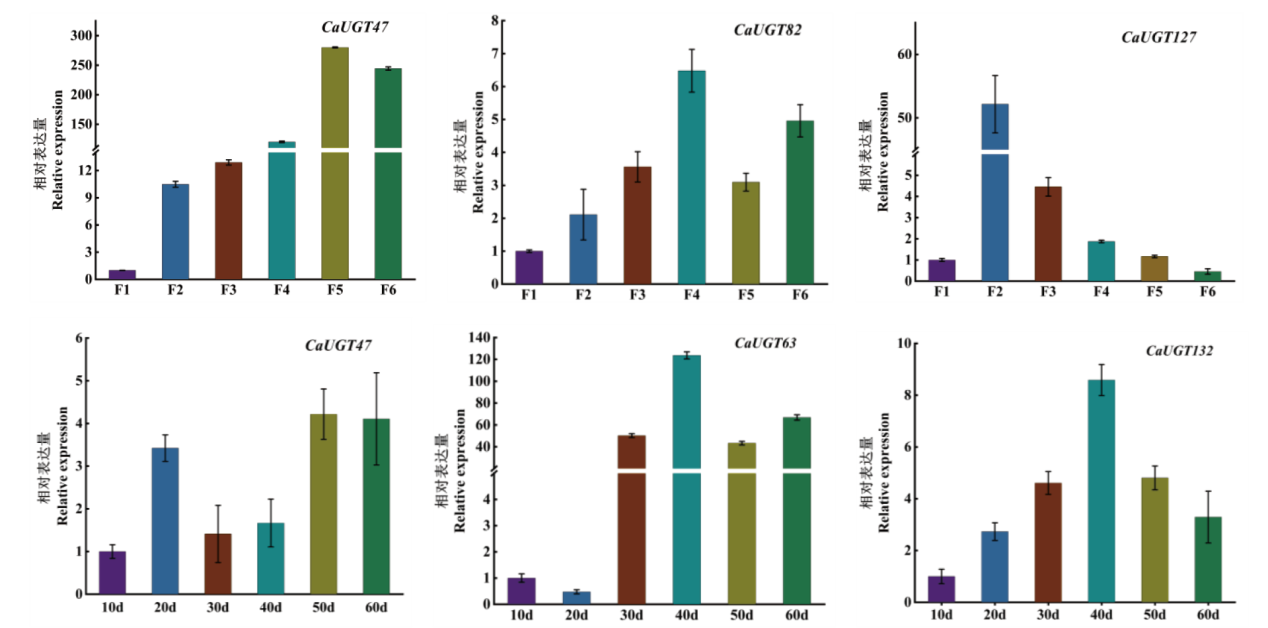
Fig. 8 Expression analysis of CaUGTs at different developmental stages of flowers and fruits F1, F2, F3, F4, F5, F5, and F6 indicate 1, 3, 5, 7, 8 and 9 d respectively
| [1] | 吴永红, 周书栋, 李雪峰, 等. 2019年辣椒科学研究进展[J]. 辣椒杂志, 2020, 18(2): 1-7, 13. |
| Wu YH, Zhou SD, Li XF, et al. The research progresses on pepper in 2019[J]. J China Capsicum, 2020, 18(2): 1-7, 13. | |
| [2] | 陶兴林, 朱惠霞, 王晓巍, 等. 5个不同产区地理标志辣椒营养成分分析[J]. 食品安全质量检测学报, 2023, 14(3): 303-310. |
| Tao XL, Zhu HX, Wang XW, et al. Nutrient composition analysis of different geographical indications peppers in 5 different production areas[J]. J Food Saf Qual, 2023, 14(3): 303-310. | |
| [3] | 王楠艺, 付文婷, 吴迪, 等. 辣椒品质研究进展[J]. 江苏农业科学, 2022, 50(16): 21-27. |
| Wang NY, Fu WT, Wu D, et al. Research progress on pepper quality[J]. Jiangsu Agric Sci, 2022, 50(16): 21-27. | |
| [4] | McKay S, Oranje P, Helin J, et al. Development of an affordable, sustainable and efficacious plant-based immunomodulatory food ingredient based on bell pepper or carrot RG-I pectic polysaccharides[J]. Nutrients, 2021, 13(3): 963. |
| [5] | 武晓艳, 叶钰怡, 任文凯, 等. 植物多糖研究进展: 功能活性及潜在机制[J]. 中国科学: 生命科学, 2023, 53(6): 808-824. |
| Wu XY, Ye YY, Ren WK, et al. Research progress of plant polysaccharides: functional activities and potential mechanisms[J]. Sci Sin Vitae, 2023, 53(6): 808-824. | |
| [6] | 刘思扬, 陆雅琦, 海日汉, 等. 功能性植物多糖及其应用研究进展[J]. 食品工业科技, 2022, 43(21): 444-453. |
| Liu SY, Lu YQ, Hai RH, et al. Research progress on plant functional polysaccharides and its application[J]. Sci Technol Food Ind, 2022, 43(21): 444-453. | |
| [7] | 吴一凡, 张睿琪, 董泽安, 等. 植物糖基转移酶的分类、分布及在果实发育中作用研究进展[J]. 食品科技, 2021, 46(7): 1-6. |
| Wu YF, Zhang RQ, Dong ZA, et al. Classification and distribution of glycosyltransferases in plant and its roles in the fruit development[J]. Food Sci Technol, 2021, 46(7): 1-6. | |
| [8] | Zhang P, Zhang Z, Zhang LJ, et al. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features[J]. Comput Struct Biotechnol J, 2020, 18: 1383-1390. |
| [9] | 陈锁英, 刘晓曼, 刘小刚, 等. 甜橙UDP-糖基转移酶基因的筛选及其功能分析[J]. 植物生理学报, 2021, 57(9): 1745-1754. |
| Chen SY, Liu XM, Liu XG, et al. Screening and functional analysis of UDP-glycosyltransferase genes in sweet orange(Citrus sinensis)[J]. Plant Physiol J, 2021, 57(9): 1745-1754. | |
| [10] | Yin QG, Shen GA, Di SK, et al. Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max[J]. Plant Cell Physiol, 2017, 58(9): 1558-1572. |
| [11] |
姚宇, 顾佳珺, 孙超, 等. 植物类黄酮UDP-糖基转移酶研究进展[J]. 生物技术通报, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
Yao Y, Gu JJ, Sun C, et al. Advances in plant flavonoids UDP-glycosyltransferase[J]. Biotechnol Bull, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
| [12] | Gharabli H, Della Gala V, Welner DH. The function of UDP-glycosyltransferases in plants and their possible use in crop protection[J]. Biotechnol Adv, 2023, 67: 108182. |
| [13] | Wu BP, Liu XH, Xu K, et al. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo(Citrus grandis)fruit[J]. BMC Plant Biol, 2020, 20(1): 459. |
| [14] | 赵倩倩, 宋艳红, 宋盼, 等. 森林草莓糖基转移酶基因家族生物信息学及其表达分析[J]. 南方农业学报, 2021, 52(6): 1615-1624. |
| Zhao QQ, Song YH, Song P, et al. Analysis of bioinformatics and gene expression of glycosyltransferase gene family in Fragaria vesca[J]. J South Agric, 2021, 52(6): 1615-1624. | |
| [15] | 祝小雅, 刘烨, 闫蕴韬, 等. 水稻UGT基因家族及成员SS4的生物信息学分析[J/OL]. 分子植物育种, 2023: 1-11. http://kns.cnki.net/kcms/detail/46.1068.S.20230505.1453.013.html. |
| Zhu XY, Liu Y, Yan YT, et al. Bioinformatics analysis of UGT gene family and its member SS4 in rice(Oryza sativa L.)[J/OL]. Molecular Plant Breeding, 2023: 1-11. http://kns.cnki.net/kcms/detail/46.1068.S.20230505.1453.013.html. | |
| [16] | 吕中睿, 刘宏, 张国昀, 等. 沙棘UGT基因家族的全基因组鉴定与表达分析[J]. 林业科学研究, 2021, 34(6): 9-19. |
| Lv ZR, Liu H, Zhang GY, et al. Genome-wide identification, characterization, and expression analysis of UGT gene family members in sea buckthorn(Hippophae rhamnoides L.)[J]. For Res, 2021, 34(6): 9-19. | |
| [17] |
Liu F, Yu HY, Deng YT, et al. PepperHub, an informatics hub for the chili pepper research community[J]. Mol Plant, 2017, 10(8): 1129-1132.
doi: S1674-2052(17)30076-X pmid: 28343897 |
| [18] |
Le Roy J, Huss B, Creach A, et al. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants[J]. Front Plant Sci, 2016, 7: 735.
doi: 10.3389/fpls.2016.00735 pmid: 27303427 |
| [19] | 唐乙钧. 糖基异构酶OsUGE3调节水稻细胞壁多糖代谢和耐盐性的机制研究[D]. 沈阳: 沈阳农业大学, 2022. |
| Tang YJ. Mechanism of OsUGE3 regulating rice cell wall polysaccharides metabolism and salt tolerance[D]. Shenyang: Shenyang Agricultural University, 2022. | |
| [20] | 陈家栋, 姜武, 宋敏全, 等. 铁皮石斛尿苷二磷酸糖基转移酶(UGT)基因家族鉴定与表达分析[J]. 中国中药杂志, 2023, 48(7): 1840-1850. |
| Chen JD, Jiang W, Song MQ, et al. Identification and expression of uridine diphosphate glycosyltransferase(UGT)gene family from Dendrobium officinale[J]. China J Chin Mater Med, 2023, 48(7): 1840-1850. | |
| [21] | Montefiori M, Espley RV, Stevenson D, et al. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit(Actinidia chinensis)[J]. Plant J, 2011, 65(1): 106-118. |
| [22] | 招雪晴, 杨静, 沈雨, 等. 石榴PgUGT基因表达特性与重组表达分析[J]. 西北植物学报, 2022, 42(3): 390-397. |
| Zhao XQ, Yang J, Shen Y, et al. Expression profiles and recombinant expression of PgUGT in pomegranate[J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(3): 390-397. | |
| [23] | Filyushin MA, Shchennikova AV, Kochieva EZ. Antocyandin-3-O-glucosyltransferase genes in pepper(Capsicum spp.) and their role in anthocyanin biosynthesis[J]. Russ J Genet, 2023, 59(5): 441-452. |
| [24] | Zheng SW, Chen ZF, Liu TT, et al. Identification and characterization of the tomato UGT gene family and effects of GAME 17 overexpression on plants and growth and development under high-CO2 conditions[J]. Agronomy, 2022, 12(9): 1998. |
| [25] | 刘蒲东, 张舒婷, 申序, 等. 龙眼UGT家族全基因组鉴定、功能预测及表达模式[J]. 应用与环境生物学报, 2021, 27(6): 1626-1635. |
| Liu PD, Zhang ST, Shen X, et al. Genome-wide identification, function prediction, and expression pattern analysis of the UGT family in Dimocarpus longan Lour[J]. Chin J Appl Environ Biol, 2021, 27(6): 1626-1635. | |
| [26] | Caputi L, Malnoy M, Goremykin V, et al. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land[J]. Plant J, 2012, 69(6): 1030-1042. |
| [27] | Zhao X, Feng Y, Ke D, et al. Molecular identification and characterization of UDP-glycosyltransferase(UGT)multigene family in Pomegranate[J]. Horticulturae, 2023, 9(5): 540-557. |
| [28] | Cui LL, Yao SB, Dai XL, et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea(Camellia sinensis)[J]. J Exp Bot, 2016, 67(8): 2285-2297. |
| [29] | Ren ZY, Ji XY, Jiao ZB, et al. Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum[J]. Hortic Res, 2020, 7: 111. |
| [30] | Rojas Rodas F, Rodriguez TO, Murai Y, et al. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside(1→6)rhamnosyltransferase[J]. Plant Mol Biol, 2014, 84(3): 287-300. |
| [31] | Dong LL, Tang ZY, Yang TY, et al. Genome-wide analysis of UGT genes in Petunia and identification of PhUGT51 involved in the regulation of salt resistance[J]. Plants, 2022, 11(18): 2434. |
| [32] | Sun LQ, Zhao LJ, Huang H, et al. Genome-wide identification, evolution and function analysis of UGTs superfamily in cotton[J]. Front Mol Biosci, 2022, 9: 965403. |
| [33] | 许永华, 卫宝瑞, 佐月, 等. 低温影响药用植物次生代谢产物积累研究进展[J]. 分子植物育种, 2022, 20(5): 1708-1715. |
| Xu YH, Wei BR, Zuo Y, et al. Research progress on the effects of low temperature on the accumulation of secondary metabolites of medicinal plants[J]. Mol Plant Breed, 2022, 20(5): 1708-1715. | |
| [34] | Javot H, Penmetsa RV, Breuillin F, et al. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis[J]. Plant J, 2011, 68(6): 954-965. |
| [35] | Zhang JX, He CM, Wu KL, et al. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis[J]. Front Plant Sci, 2016, 7: 5. |
| [36] | Zhang GQ, Xu Q, Bian C, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution[J]. Sci Rep, 2016, 6: 19029. |
| [1] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| [2] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [3] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [4] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [5] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [6] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [7] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [8] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [9] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [10] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [11] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [12] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [13] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [14] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [15] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||