








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 181-189.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0080
Previous Articles Next Articles
DU Wei( ), LI Zhi-min(
), LI Zhi-min( ), XING Yan-ming, LIU Pu-lin, MIAO Li-hong
), XING Yan-ming, LIU Pu-lin, MIAO Li-hong
Received:2024-01-19
Online:2024-09-26
Published:2024-10-12
Contact:
LI Zhi-min
E-mail:604003648@qq.com;lizhimin1119@163.com
DU Wei, LI Zhi-min, XING Yan-ming, LIU Pu-lin, MIAO Li-hong. Screening and Identification of a Bacillus licheniformis Strain with High Electro-transfection Efficiency and Elevated Biomass[J]. Biotechnology Bulletin, 2024, 40(9): 181-189.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| gyrB-F[ | GCCGGCTTCATGGGTTCCG |
| gyrB-R[ | GCGTCGGTGCTTCTGTTG |
| 27f[ | AGAGTTTGATCCTGGCTCAG |
| 1492r[ | GGTTACCTTGTTACGACTT |
| GFP-F | CTGCGGCCGGTGCACATATGATGGTGAGCAAGGGCGAG |
| GFP-R | CTGCAGGTCGACAAGCTTCTACTTGTACAGCTCGTCCATG |
| pBES-F | AAGCTTGTCGACCTGCAGTC |
| pBES-R | CATATGCACCGGCCGC |
Table 1 Primer sequences used in this experiment
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| gyrB-F[ | GCCGGCTTCATGGGTTCCG |
| gyrB-R[ | GCGTCGGTGCTTCTGTTG |
| 27f[ | AGAGTTTGATCCTGGCTCAG |
| 1492r[ | GGTTACCTTGTTACGACTT |
| GFP-F | CTGCGGCCGGTGCACATATGATGGTGAGCAAGGGCGAG |
| GFP-R | CTGCAGGTCGACAAGCTTCTACTTGTACAGCTCGTCCATG |
| pBES-F | AAGCTTGTCGACCTGCAGTC |
| pBES-R | CATATGCACCGGCCGC |
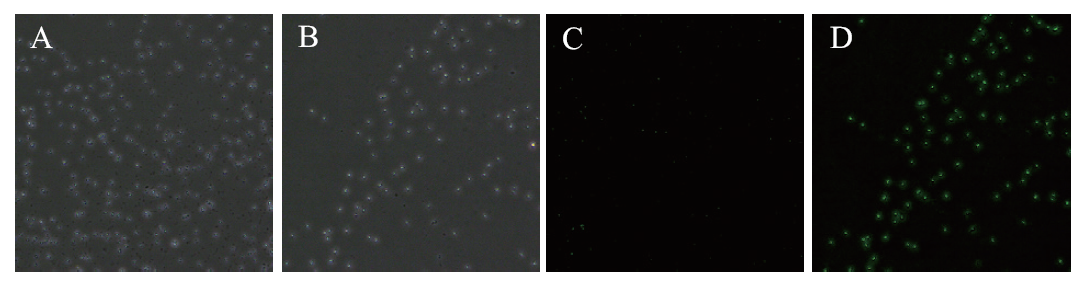
Fig. 2 Fluorescence microscope observation of green fluorescence of bacteriophage A: Bright field observation of wild strains 1-33. B: Bright field observation of strains 1-33 transfected with pBES-P43-egfp. C: Fluorescence-excited observation of wild strains 1-33. D: Fluorescence-excited observation of strains 1-33 transferred to pBES-P43-egfp
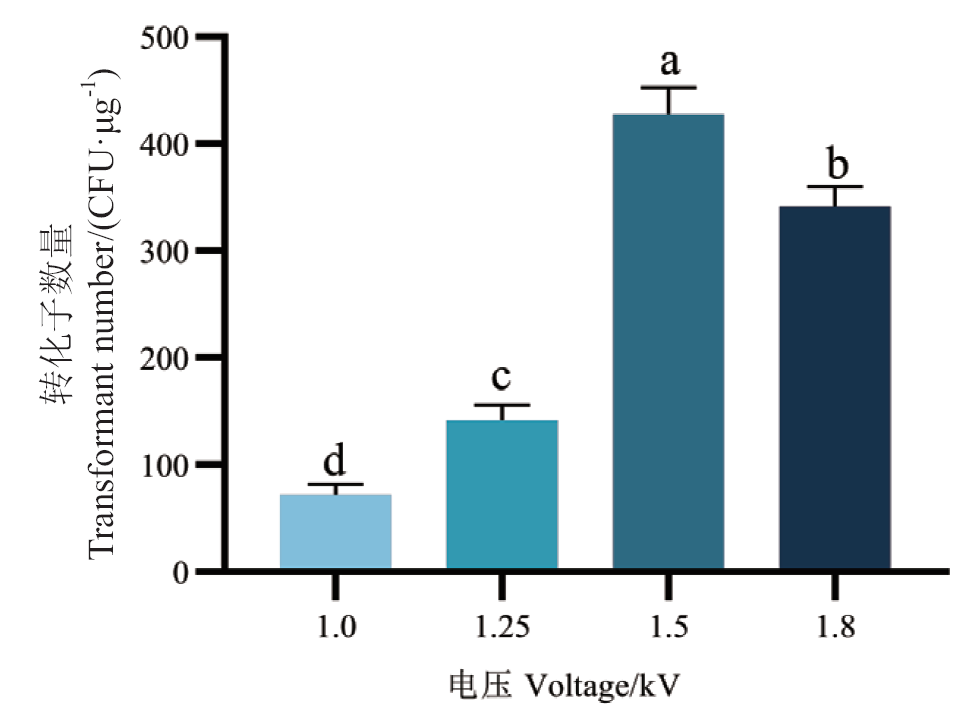
Fig. 3 Effects of different voltages on the electro-transfection efficiency of B. licheniformis 2709 Different lowercase letters indicate significant differences(P < 0.05), the same below
| 菌株编号 Strain No. | 转化效率Electro-transfection efficiency/ (CFU·μg-1 DNA) | 菌株编号 Strain No. | 转化效率Electro-transfection efficiency/(CFU·μg-1 DNA) | |
|---|---|---|---|---|
| 1-4 | 1 420±20 | 1-25 | 1 570±81 | |
| 1-5 | 3 650±61 | 1-27 | 855±56 | |
| 1-8 | 0±0 | 1-33 | 6 700±89 | |
| 1-16 | 270±22 | 1-37 | 260±35 | |
| 1-17 | 118±9 | 2709 | 490±29 | |
| 1-19 | 416±46 |
Table 2 Electro-transfection efficiency of different strains
| 菌株编号 Strain No. | 转化效率Electro-transfection efficiency/ (CFU·μg-1 DNA) | 菌株编号 Strain No. | 转化效率Electro-transfection efficiency/(CFU·μg-1 DNA) | |
|---|---|---|---|---|
| 1-4 | 1 420±20 | 1-25 | 1 570±81 | |
| 1-5 | 3 650±61 | 1-27 | 855±56 | |
| 1-8 | 0±0 | 1-33 | 6 700±89 | |
| 1-16 | 270±22 | 1-37 | 260±35 | |
| 1-17 | 118±9 | 2709 | 490±29 | |
| 1-19 | 416±46 |
| 菌株 Strain | 湿重 Wet weight/(g·L-1) | 干重 Dry weight/(g·L-1) |
|---|---|---|
| 1-33 | 42.8±1.05 | 6.8±0.74 |
| B. licheniformis 2709 | 33.1±0.42 | 5.2±0.11 |
Table 3 Comparison of wet and dry weights of strain 1-33 and B. licheniformis 2709
| 菌株 Strain | 湿重 Wet weight/(g·L-1) | 干重 Dry weight/(g·L-1) |
|---|---|---|
| 1-33 | 42.8±1.05 | 6.8±0.74 |
| B. licheniformis 2709 | 33.1±0.42 | 5.2±0.11 |
| 实验项目 Experimental project | 结果 Result | 实验项目 Experimental project | 结果 Result | |
|---|---|---|---|---|
| 利用木糖 | + | 厌氧生长 | + | |
| 利用阿拉伯糖 | + | V-P实验 | + | |
| 甘露醇 | + | pH=5.7生长 | + | |
| 明胶液化 | + | 柠檬酸盐 | + | |
| 7% NaCl生长 | + | 丙酸盐 | - | |
| 淀粉水解 | + | 硝酸盐还原 | + |
Table 4 Physiological and biochemical characterization of strain 1-33
| 实验项目 Experimental project | 结果 Result | 实验项目 Experimental project | 结果 Result | |
|---|---|---|---|---|
| 利用木糖 | + | 厌氧生长 | + | |
| 利用阿拉伯糖 | + | V-P实验 | + | |
| 甘露醇 | + | pH=5.7生长 | + | |
| 明胶液化 | + | 柠檬酸盐 | + | |
| 7% NaCl生长 | + | 丙酸盐 | - | |
| 淀粉水解 | + | 硝酸盐还原 | + |
| 抗生素种类 Antibiotic type | 抑菌浓度 Inhibitory concentration/(μg·mL-1) |
|---|---|
| 氨苄青霉素Benzylpenicillin | 10 |
| 四环素Tetracycline | 25 |
| 卡那霉素Kanamycin | 5 |
| 红霉素Erythromycin | 2 |
Table 5 Inhibitory concentrations of four antibiotics against B. licheniformis 1-33
| 抗生素种类 Antibiotic type | 抑菌浓度 Inhibitory concentration/(μg·mL-1) |
|---|---|
| 氨苄青霉素Benzylpenicillin | 10 |
| 四环素Tetracycline | 25 |
| 卡那霉素Kanamycin | 5 |
| 红霉素Erythromycin | 2 |
| [1] | Chen JQ, Zhu YM, Fu G, et al. High-level intra- and extra-cellular production of D-psicose 3-epimerase via a modified xylose-inducible expression system in Bacillus subtilis[J]. J Ind Microbiol Biotechnol, 2016, 43(11): 1577-1591. |
| [2] | 张莹, 韩晓静, 蔡逸安, 等. 解淀粉芽孢杆菌内源启动子的筛选及表达碱性果胶酶的应用研究[J]. 微生物学报, 2023, 63(4): 1575-1586. |
| Zhang Y, Han XJ, Cai YA, et al. Screening of endogenous promoters of Bacillus amyloliquefaciens and application of them in the expression of alkaline pectinase[J]. Acta Microbiol Sin, 2023, 63(4): 1575-1586. | |
| [3] | Shrestha S, Chio C, Khatiwada JR, et al. Optimization of multiple enzymes production by fermentation using lipid-producing Bacillus sp[J]. Front Microbiol, 2022, 13: 1049692. |
| [4] | Elemosho R, Suwanto A, Thenawidjaja M. Extracellular expression in Bacillus subtilis of a thermostable Geobacillus stearothermophilus lipase[J]. Electron J Biotechnol, 2021, 53: 71-79. |
| [5] |
Wang Q, Zheng H, Wan X, et al. Optimization of inexpensive agricultural by-products as raw materials for bacitracin production in Bacillus licheniformis DW2[J]. Appl Biochem Biotechnol, 2017, 183(4): 1146-1157.
doi: 10.1007/s12010-017-2489-1 pmid: 28593603 |
| [6] | Li YX, Li ZR, Yamanaka K, et al. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis[J]. Sci Rep, 2015, 5: 9383. |
| [7] | He FM, Gao BJ, Cheng X, et al. High-level production of poly-γ-glutamic acid by a newly isolated Bacillus sp. YJY-8 and potential use in increasing the production of tomato[J]. Prep Biochem Biotechnol, 2024, 54(5): 637-646. |
| [8] |
Cai D, Rao Y, Zhan Y, et al. Engineering Bacillus for efficient production of heterologous protein: current progress, challenge and prospect[J]. J Appl Microbiol, 2019, 126(6): 1632-1642.
doi: 10.1111/jam.14192 pmid: 30609144 |
| [9] | Souza CC, Guimarães JM, Pereira SDS, et al. The multifunctionality of expression systems in Bacillus subtilis: emerging devices for the production of recombinant proteins[J]. Exp Biol Med, 2021, 246(23): 2443-2453. |
| [10] | Liu HL, Wang S, Song LX, et al. Trehalose production using recombinant trehalose synthase in Bacillus subtilis by integrating fermentation and biocatalysis[J]. J Agric Food Chem, 2019, 67(33): 9314-9324. |
| [11] |
Zhang K, Su LQ, Wu J. Enhanced extracellular pullulanase production in Bacillus subtilis using protease-deficient strains and optimal feeding[J]. Appl Microbiol Biotechnol, 2018, 102(12): 5089-5103.
doi: 10.1007/s00253-018-8965-x pmid: 29675805 |
| [12] |
Wang Y, Chen ZM, Zhao RL, et al. Deleting multiple lytic genes enhances biomass yield and production of recombinant proteins by Bacillus subtilis[J]. Microb Cell Fact, 2014, 13: 129.
doi: 10.1186/s12934-014-0129-9 pmid: 25176138 |
| [13] | Wang Q, Yu HM, Wang MM, et al. Enhanced biosynthesis and characterization of surfactin isoforms with engineered Bacillus subtilis through promoter replacement and Vitreoscilla hemoglobin co-expression[J]. Process Biochemistry, 2018, 70:36-44. |
| [14] | Muras A, Romero M, Mayer C, et al. Biotechnological applications of Bacillus licheniformis[J]. Crit Rev Biotechnol, 2021, 41(4): 609-627. |
| [15] | Zhou CX, Zhou HY, Zhang HT, et al. Optimization of alkaline protease production by rational deletion of sporulation related genes in Bacillus licheniformis[J]. Microb Cell Fact, 2019, 18(1): 127. |
| [16] | Shen PL, Niu DD, Liu XL, et al. High-efficiency chromosomal integrative amplification strategy for overexpressing α-amylase in Bacillus licheniformis[J]. J Ind Microbiol Biotechnol, 2022, 49(3): kuac009. |
| [17] | 陈坤. 2709碱性蛋白酶的高产工程菌株构建及应用性能分析[D]. 天津: 天津科技大学, 2018. |
| Chen K. Construction of the engineering bacteria with high yield of alkaline protease 2709 and its application performance[D]. Tianjin:Tianjin University of Science & Technology, 2018. | |
| [18] | Xue GP, Johnson JS, Dalrymple BP. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis[J]. J Microbiol Meth, 1999, 34(3): 183-191. |
| [19] | Huang CH, Chang MT, Huang LN, et al. Development of a novel PCR assay based on the gyrase B gene for species identification of Bacillus licheniformis[J]. Mol Cell Probes, 2012, 26(5): 215-217. |
| [20] | 马凯, 刘光全, 程池. 地衣芽孢杆菌16S rRNA基因的TD-PCR扩增及系统发育分析[J]. 微生物学通报, 2007, 34(4): 709-711. |
| Ma K, Liu GQ, Cheng C. The TD-PCR and phylogenetic analysis of Bacillus licheniformis 16S rDNA[J]. Microbiology, 2007, 34(4): 709-711. | |
| [21] | 东秀珠, 蔡妙英. 常见细菌系统鉴定手册[M]. 北京: 科学出版社, 2001. |
| Dong XZ, Cai MY. Handbook of identification of common bacterial systems[M]. Beijing: Science Press, 2001. | |
| [22] |
Brockmeier U, Caspers M, Freudl R, et al. Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria[J]. J Mol Biol, 2006, 362(3): 393-402.
pmid: 16930615 |
| [23] | Zhan YY, Xu Y, Zheng PL, et al. Establishment and application of multiplexed CRISPR interference system in Bacillus licheniformis[J]. Appl Microbiol Biotechnol, 2020, 104(1): 391-403. |
| [24] |
Zakataeva NP, Nikitina OV, Gronskiy SV, et al. A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains[J]. Appl Microbiol Biotechnol, 2010, 85(4): 1201-1209.
doi: 10.1007/s00253-009-2276-1 pmid: 19820923 |
| [25] | He HH, Zhang YP, Shi GY, et al. Recent biotechnological advances and future prospective of Bacillus licheniformis as microbial cell factories[J]. Syst Microbiol Biomanuf, 2023, 3(4): 521-532. |
| [26] | Ahn S, Jun SM, Ro HJ, et al. Complete genome of Bacillus subtilis subsp. subtilis KCTC 3135T and variation in cell wall genes of B. subtilis strains[J]. J Microbiol Biotechnol, 2018, 28(10): 1760-1768. |
| [27] | Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis[J]. Nature, 1997, 390(6657): 249-256. |
| [28] | Rahimnahal S, Meimandipour A, Fayazi J, et al. Biochemical and molecular characterization of novel keratinolytic protease from Bacillus licheniformis(KRLr1)[J]. Front Microbiol, 2023, 14: 1132760. |
| [29] |
Priya I, Dhar MK, Bajaj BK, et al. Cellulolytic activity of thermophilic bacilli isolated from tattapani hot spring sediment in North West Himalayas[J]. Indian J Microbiol, 2016, 56(2): 228-231.
doi: 10.1007/s12088-016-0578-4 pmid: 27570317 |
| [30] | 全爽, 陈涛, 毛宗林, 等. 一株土壤源地衣芽孢杆菌的鉴定及生物学特性研究[J]. 中国畜牧杂志, 2023, 59(1): 217-222. |
| Quan S, Chen T, Mao ZL, et al. Identification and biological characterization of a soil-derived Bacillus licheniformis strain[J]. Chin J Anim Sci, 2023, 59(1): 217-222. | |
| [31] | Li YR, Wang HR, Zhang L, et al. Efficient genome editing in Bacillus licheniformis mediated by a conditional CRISPR/Cas9 system[J]. Microorganisms, 2020, 8(5): 754. |
| [32] | Hoffmann K, Wollherr A, Larsen M, et al. Facilitation of direct conditional knockout of essential genes in Bacillus licheniformis DSM13 by comparative genetic analysis and manipulation of genetic competence[J]. Appl Environ Microbiol, 2010, 76(15): 5046-5057. |
| [33] | 莫静燕, 陈献忠, 王正祥. 地衣芽孢杆菌原生质体的制备、再生及转化研究[J]. 生物技术, 2009, 19(5): 75-77. |
| Mo JY, Chen XZ, Wang ZX. Preparation, regeneration and genetic transformation of Bacillus licheniformis protoplasts[J]. Biotechnology, 2009, 19(5): 75-77. | |
| [34] | Zhang Y, Hu JM, Zhang Q, et al. Enhancement of alkaline protease production in recombinant Bacillus licheniformis by response surface methodology[J]. Bioresour Bioprocess, 2023, 10(1): 27. |
| [35] | 肖静, 张虎, 李子源, 等. 抑制菌体自溶的地衣芽孢杆菌工程菌及其构建方法和应用:CN109868253A[P]. 2019-06-11. |
| Xiao J, Zhang H, Li ZY, et al. Bacillus licheniformis engineered bacteria that inhibit bacterial autolysis and its construction method and application: CN109868253A[P]. 2019-06-11. |
| [1] | ZHANG Ya-ya, LI Pan-pan, GAO Hui-hui, JIA Chen-bo, XU Chun-yan. Exploring on the Pathogenesis of Root Rot of Lycium barbarum cv. ‘Ningqi-5' Based on the Rhizoplane Fungal Community and Pathogens Identification [J]. Biotechnology Bulletin, 2024, 40(9): 238-248. |
| [2] | YUAN Lan, HUANG Ya-nan, ZHANG Bei-ni, XIONG Yu-meng, WANG Hong-yang. High-throughput Sample Preparation Method for the Identification of Potato Ploidy Using Flow Cytometry [J]. Biotechnology Bulletin, 2024, 40(9): 141-147. |
| [3] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [4] | TAN Bo-wen, ZHANG Yi, ZHANG Peng, WANG Zhen-yu, MA Qiu-xiang. Identification and Bioinformatics Analysis of Gene in the Magnesium Transporter Family in Cassava [J]. Biotechnology Bulletin, 2024, 40(9): 20-32. |
| [5] | XING Li-nan, ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen. Analysis of DoWRKY40 Gene Expression Characteristics and Screening of Interacting Proteins in Yam [J]. Biotechnology Bulletin, 2024, 40(8): 118-128. |
| [6] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [7] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [8] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [9] | YANG Lu, YUAN Yuan, FANG Zhi-kai, LIN Ru, JIANG Hong, ZHOU Jian. Identification of a Streptomyces Strain and Study on the Fermentation Process of Geldanamycin Production [J]. Biotechnology Bulletin, 2024, 40(6): 299-309. |
| [10] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [11] | XU Wei-fang, LI He-yu, ZHANG Hui, HE Zi-ang, GAO Wen-heng, XIE Zi-yang, WANG Chuan-wen, YIN Deng-ke. Efficacy and Its Mechanism of Bacterial Strain HX0037 on the Control of Anthracnose Disease of Trichosanthes kirilowii Maxim [J]. Biotechnology Bulletin, 2024, 40(4): 228-241. |
| [12] | RAO Yong-bin, ZHANG Jun-li. Biological Characteristics, Domestication and Cultivation of Wild Gymnopilus subpurpuratus [J]. Biotechnology Bulletin, 2024, 40(4): 264-270. |
| [13] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [14] | WANG Lu, LIU Meng-yu, ZHANG Fu-yuan, JI Shou-kun, WANG Yun, ZHANG Ying-jie, DUAN Chun-hui, LIU Yue-qin, YAN Hui. Isolation and Identification of Rumen Skatole-degrading Bacteria and Analysis on Their Degradation Characteristics [J]. Biotechnology Bulletin, 2024, 40(3): 305-311. |
| [15] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||