








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 176-187.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0903
WANG Tian-xi( ), YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia(
), YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia( )
)
Received:2024-09-19
Online:2025-04-26
Published:2025-04-25
Contact:
LIANG Mei-xia
E-mail:2864509389@qq.com;mxliangdd@163.com
WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene[J]. Biotechnology Bulletin, 2025, 41(4): 176-187.
| 基因ID Gene ID | 基因名称 Gene name | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位预测 Localization prediction |
|---|---|---|---|---|
| MD00G1059100 | MdPLATZ1 | 8.55 | 28 544.59 | Nucleus |
| MD02G1017000 | MdPLATZ2 | 8.56 | 29 394.72 | Nucleus |
| MD02G1208800 | MdPLATZ3 | 8.82 | 24 671.44 | Nucleus |
| MD03G1129200 | MdPLATZ4 | 9.51 | 21 971.14 | Nucleus |
| MD05G1248500 | MdPLATZ5 | 8.39 | 28 487.59 | Chloroplast |
| MD06G1001300 | MdPLATZ6 | 8.78 | 28 441.46 | Chloroplast, nucleus |
| MD06G1035500 | MdPLATZ7 | 9.25 | 24 878.17 | Nucleus |
| MD07G1117000 | MdPLATZ8 | 8.81 | 27 385.73 | Nucleus |
| MD10G1229000 | MdPLATZ9 | 8.39 | 28 548.56 | Chloroplast, nucleus |
| MD11G1151300 | MdPLATZ10 | 9.30 | 24 312.98 | Nucleus |
| MD12G1103000 | MdPLATZ11 | 8.88 | 22 043.09 | Nucleus |
| MD13G1017800 | MdPLATZ12 | 9.50 | 24 708.19 | Nucleus |
| MD14G1097400 | MdPLATZ13 | 8.52 | 21 996.09 | Nucleus |
| MD15G1161500 | MdPLATZ14 | 8.75 | 29 101.53 | Nucleus |
| MD16G1015800 | MdPLATZ15 | 9.02 | 25 758.81 | Nucleus |
| MD16G1273400 | MdPLATZ16 | 9.33 | 25 264.61 | Nucleus |
| MD17G1254800 | MdPLATZ17 | 8.24 | 17 230.53 | Nucleus |
Table 1 Information of MdPLATZ gene family member in apple
| 基因ID Gene ID | 基因名称 Gene name | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位预测 Localization prediction |
|---|---|---|---|---|
| MD00G1059100 | MdPLATZ1 | 8.55 | 28 544.59 | Nucleus |
| MD02G1017000 | MdPLATZ2 | 8.56 | 29 394.72 | Nucleus |
| MD02G1208800 | MdPLATZ3 | 8.82 | 24 671.44 | Nucleus |
| MD03G1129200 | MdPLATZ4 | 9.51 | 21 971.14 | Nucleus |
| MD05G1248500 | MdPLATZ5 | 8.39 | 28 487.59 | Chloroplast |
| MD06G1001300 | MdPLATZ6 | 8.78 | 28 441.46 | Chloroplast, nucleus |
| MD06G1035500 | MdPLATZ7 | 9.25 | 24 878.17 | Nucleus |
| MD07G1117000 | MdPLATZ8 | 8.81 | 27 385.73 | Nucleus |
| MD10G1229000 | MdPLATZ9 | 8.39 | 28 548.56 | Chloroplast, nucleus |
| MD11G1151300 | MdPLATZ10 | 9.30 | 24 312.98 | Nucleus |
| MD12G1103000 | MdPLATZ11 | 8.88 | 22 043.09 | Nucleus |
| MD13G1017800 | MdPLATZ12 | 9.50 | 24 708.19 | Nucleus |
| MD14G1097400 | MdPLATZ13 | 8.52 | 21 996.09 | Nucleus |
| MD15G1161500 | MdPLATZ14 | 8.75 | 29 101.53 | Nucleus |
| MD16G1015800 | MdPLATZ15 | 9.02 | 25 758.81 | Nucleus |
| MD16G1273400 | MdPLATZ16 | 9.33 | 25 264.61 | Nucleus |
| MD17G1254800 | MdPLATZ17 | 8.24 | 17 230.53 | Nucleus |

Fig. 2 Homology analysis of PLATZ genes in appleGrey curves indicate regions of colinearity in the apple genome, and red curves indicate pairs of genes where segmental duplication occurs

Fig. 3 PLATZ phylogenetic tree of apple, Arabidopsis and maize▲ indicates apple's PLATZ protein. ★ indicates maize's PLATZ protein. ● indicates Arabidopsis's PLATZprotein. Numbers indicate the closeness of kinship

Fig. 4 Expressions of MdPLATZs gene in different tissues of appleWS: Whole seedling; FR: fruit; L: leaf; SA: stem tip; FL: flower; S: stem; EDB: early dormant bud; FDB: bloom bud; YL: young

Fig. 6 Evolutionary relationship between MdPLATZs and ORESARA15The numbers in the figure indicate the proximity of the relatives and the branch lengths, respectively
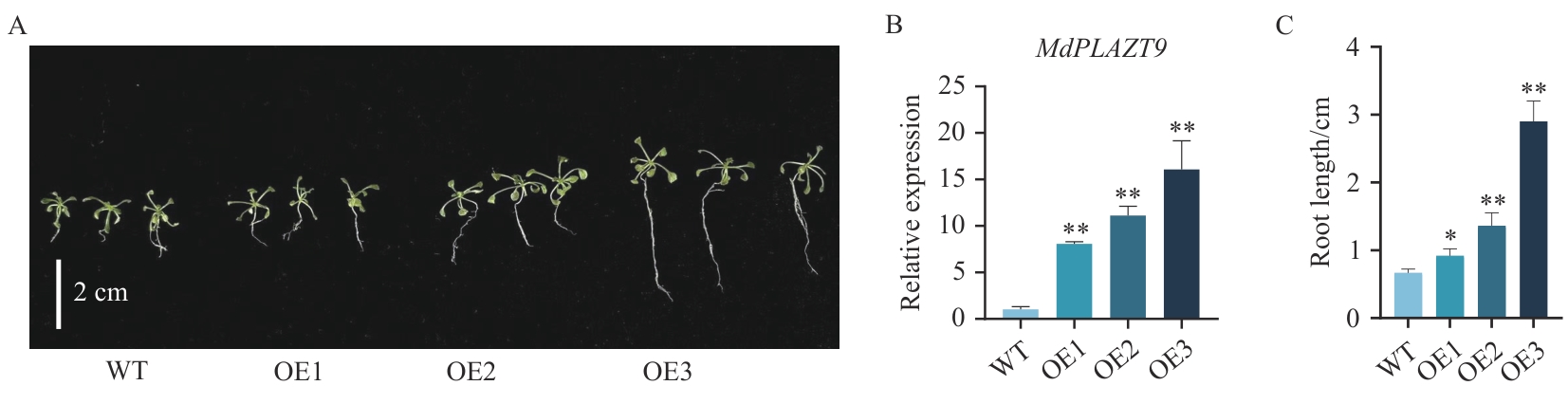
Fig. 7 Comparison of transgenic A. thaliana with wild-type A. thalianaA: Phenotypes of wild-type Arabidopsisthaliana and overexpressed A. thaliana grown for 14 d. B: Expression of MdPLATZ9 in wild-type A. thaliana and overexpressed A. thaliana. C: Statistics of root length data of wild-type A. thaliana and overexpressed A. thaliana grown for 14 d. WT: Wild-type A. thaliana; OE1: overexpressed A. thaliana line; OE2: overexpressed line 2; OE3: overexpressed line 3; the same below. The data are means±standard deviation (SD), n=3. * P<0.05; ** P<0.01, *** P<0.001

Fig. 8 Determination of phenotypic and physiological parameters of wild-type and overexpressed strains of A. thaliana under drought stressA: Phenotypes of wild-type A. thaliana and overexpressed A. thaliana under drought stress. B‒G: Rate of O2- production, H2O2 content, CAT activity, POD activity, Vc activity, and MDA content of wild-type A. thaliana and overexpressed A. thaliana under drought stress. Data are the mean±SD of three replicates. Different letters indicate significant differences at P<0.05 level. The same below
| 1 | Strader L, Weijers D, Wagner D. Plant transcription factors-being in the right place with the right company [J]. Curr Opin Plant Biol, 2022, 65: 102136. |
| 2 | Zhang KM, Lan YG, Shi YN, et al. Systematic analysis and functional characterization of the PLATZ transcription factors in moso bamboo (Phyllostachys edulis) [J]. J Plant Growth Regul, 2023, 42(1): 218-236. |
| 3 | Nagano Y, Furuhashi H, Inaba T, et al. A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences [J]. Nucleic Acids Res, 2001, 29(20): 4097-4105. |
| 4 | González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, et al. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana [J]. Proc Natl Acad Sci U S A, 2016, 113(35): E5232-E5241. |
| 5 | Kim JH, Kim J, Jun SE, et al. ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis [J]. New Phytol, 2018, 220(2): 609-623. |
| 6 | Dong T, Yin XM, Wang HT, et al. ABA-INDUCED expression 1 is involved in ABA-inhibited primary root elongation via modulating ROS homeostasis in Arabidopsis [J]. Plant Sci, 2021, 304: 110821. |
| 7 | Li Q, Wang JC, Ye JW, et al. The maize imprinted gene floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III [J]. Plant Cell, 2017, 29(10): 2661-2675. |
| 8 | Li H, Wang YY, Xiao QL, et al. Transcription factor ZmPLATZ2 positively regulate the starch synthesis in maize [J]. Plant Growth Regul, 2021, 93(3): 291-302. |
| 9 | Wang AH, Hou QQ, Si LZ, et al. The PLATZ transcription factor GL6 affects grain length and number in rice [J]. Plant Physiol, 2019, 180(4): 2077-2090. |
| 10 | Zhou SR, Xue HW. The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division [J]. J Integr Plant Biol, 2020, 62(6): 847-864. |
| 11 | Azim JB, Khan MFH, Hassan L, et al. Genome-wide characterization and expression profiling of plant-specific PLATZ transcription factor family genes in Brassica rapa L. [J]. Plant Breed Biotech, 2020, 8(1): 28-45. |
| 12 | Wang JC, Ji C, Li Q, et al. Genome-wide analysis of the plant-specific PLATZ proteins in maize and identification of their general role in interaction with RNA polymerase III complex [J]. BMC Plant Biol, 2018, 18(1): 221. |
| 13 | Fu YX, Cheng MP, Li ML, et al. Identification and characterization of PLATZ transcription factors in wheat [J]. Int J Mol Sci, 2020, 21(23): 8934. |
| 14 | Zhang SC, Yang R, Huo YQ, et al. Expression of cotton PLATZ1 in transgenic Arabidopsis reduces sensitivity to osmotic and salt stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways [J]. BMC Plant Biol, 2018, 18(1): 218. |
| 15 | Blair EJ, Bonnot T, Hummel M, et al. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis [J]. Sci Rep, 2019, 9(1): 4814. |
| 16 | So H, Choi SJ, Chung E, et al. Molecular characterization of stress-inducible PLATZ gene from soybean (Glycine max L.) [J]. Plant Omics, 2015, 8(6):479-484. |
| 17 | Liu SS, Yang R, Liu M, et al. PLATZ2 negatively regulates salt tolerance in Arabidopsis seedlings by directly suppressing the expression of the CBL4/SOS3 and CBL10/SCaBP8 genes [J]. J Exp Bot, 2020, 71(18): 5589-5602. |
| 18 | Zenda T, Liu ST, Wang X, et al. Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting inbred lines [J]. Int J Mol Sci, 2019, 20(6): 1268. |
| 19 | Zhao JY, Zheng L, Wei JT, et al. The soybean PLATZ transcription factor GmPLATZ17 suppresses drought tolerance by interfering with stress-associated gene regulation of GmDREB5 [J]. Crop J, 2022, 10(4): 1014-1025. |
| 20 | Qi JH, Wang H, Wu XY, et al. Genome-wide characterization of the PLATZ gene family in watermelon (Citrullus lanatus L.) with putative functions in biotic and abiotic stress response [J]. Plant Physiol Biochem, 2023, 201: 107854. |
| 21 | Starkus A, Morkūnaitė-Haimi Š, Gurskas T, et al. The biological and genetic mechanisms of fruit drop in apple tree (Malus × domestica Borkh.) [J]. Horticulturae, 2024, 10(9): 987. |
| 22 | Urrestarazu J, Muranty H, Denancé C, et al. Genome-wide association mapping of flowering and ripening periods in apple [J]. Front Plant Sci, 2017, 8: 1923. |
| 23 | Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0 [J]. Mol Biol Evol, 2013, 30(12): 2725-2729. |
| 24 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 25 | Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J, 1998, 16(6): 735-743. |
| 26 | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method [J]. Nat Protoc, 2008, 3: 1101-1108. |
| 27 | Meng D, Li YY, Bai Y, et al. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress [J]. Plant Physiol Biochem, 2016, 103: 71-83. |
| 28 | Huang C, Yu QB, Lv RH, et al. The reduced plastid-encoded polymerase-dependent plastid gene expression leads to the delayed greening of the Arabidopsis fln2 mutant [J]. PLoS One, 2013, 8(9): e73092. |
| 29 | Farinati S, Rasori A, Varotto S, et al. Rosaceae fruit development, ripening and post-harvest: an epigenetic perspective [J]. Front Plant Sci, 2017, 8: 1247. |
| 30 | Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.) [J]. Nat Genet, 2010, 42(10): 833-839. |
| 31 | Han XL, Liu K, Yuan GP, et al. Genome-wide identification and characterization of AINTEGUMENTA-LIKE (AIL) family genes in apple (Malus domestica Borkh.) [J]. Genomics, 2022, 114(2): 110313. |
| 32 | Meyers BC, Kozik A, Griego A, et al. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis [J]. Plant Cell, 2003, 15(4): 809-834. |
| 33 | Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome [J]. Science, 2001, 291(5507): 1304-1351. |
| 34 | Timilsina R, Kim Y, Park S, et al. ORESARA15, a PLATZ transcription factor, controls root meristem size through auxin and cytokinin signalling-related pathways [J]. J Exp Bot, 2022, 73(8): 2511-2524. |
| 35 | Cai KF, Song XJ, Yue WH, et al. Identification and functional characterization of abiotic stress tolerance-related PLATZ transcription factor family in barley (Hordeum vulgare L.) [J]. Int J Mol Sci, 2024, 25(18): 10191. |
| 36 | Zhang KM, Lan YG, Wu M, et al. PhePLATZ1, a PLATZ transcription factor in moso bamboo (Phyllostachys edulis), improves drought resistance of transgenic Arabidopsis thaliana [J]. Plant Physiol Biochem, 2022, 186: 121-134. |
| 37 | Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana [J]. Plant J, 2005, 43(1): 68-78. |
| 38 | 冯军, 郑彩霞. DREB转录因子在植物非生物胁迫中的作用及应用研究 [J]. 植物生理学报, 2011, 47(5): 437-442. |
| Feng J, Zheng CX. Research and application prospect of DREB transcription factor in plant abiotic stress resistance [J]. Plant Physiol J, 2011, 47(5): 437-442. | |
| 39 | Chen YF, Li Z, Sun TT, et al. Sugarcane ScDREB2B-1 confers drought stress tolerance in transgenic Nicotiana benthamiana by regulating the ABA signal, ROS level and stress-related gene expression[J]. Int J Mol Sci,2022, 23(17): 9557. |
| [1] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [2] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [3] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [4] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [5] | WANG Chen, LIU Guo-mei, CHEN Chang, ZHANG Jin-long, YAO Lin, SUN Xuan, DU Chun-fang. Genome-wide Identification and Expression Analysis of CCDs Family in Brassia rapa L. [J]. Biotechnology Bulletin, 2025, 41(3): 161-170. |
| [6] | HAN Jiang-tao, ZHANG Shuai-bo, QIN Ya-rui, HAN Shuo-yang, ZHANG Ya-kang, WANG Ji-qing, DU Qing-jie, XIAO Huai-juan, LI Meng. Identification of β-amylase Gene Family in Melon and Their Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(3): 171-180. |
| [7] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [8] | WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(3): 255-270. |
| [9] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [10] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [11] | YAN Wei, CHEN Hui-ting, YE Qing, LIU Guang-chao, LIU Xin, HOU Li-xia. Identification of the Grape HCT Gene Family and Their Responses to Low-temperature Stress [J]. Biotechnology Bulletin, 2025, 41(2): 175-186. |
| [12] | LI Yan-wei, YANG Yan-yan, SUN Ya-ling, HUO Yu-meng, WANG Zhen-bao, LIU Bing-jiang. Regulation Mechanism of Plant Hormones Related to Onion Bulb Enlargement and Development Based on Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 187-201. |
| [13] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [14] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| [15] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||