








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 208-217.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0019
GUO Tao( ), AI Li-jiao(
), AI Li-jiao( ), ZOU Shi-hui, ZHOU Ling, LI Xue-mei
), ZOU Shi-hui, ZHOU Ling, LI Xue-mei
Received:2025-01-08
Online:2025-06-26
Published:2025-06-30
Contact:
AI Li-jiao
E-mail:yushen0002008@126.com;alj01461@foxmail.com
GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay[J]. Biotechnology Bulletin, 2025, 41(6): 208-217.
| 名称 Primer name | 序列 Primer sequence (5′‒3′) |
|---|---|
| RAV1-f | ATGGATGGTAGTTACATAGATGAGA |
| RAV1-r | TTACAGAGCCCCAATTATCCTTTGT |
| 3302-R1-1 | GAACACGGGGGACTCTTGACATGGATGGTAGTTACATAGA |
| 3302-R1-2 | TTTACCCTCAGATCTACCATCAGAGCCCCAATTATCCTTT |
| RAV1-RT-1 | TAAAGGTTGGAGCCGATTTGTGA |
| RAV1-RT-2 | ACCCGTTTCTCGTATTCCAGTCA |
| CjGAPDH-1 | GGGAATCCTTGGTTACACTGAG |
| CjGAPDH-2 | ACCCCATTCGTTGTCATACC |
| Atactin-1 | ACCACTGTCCACTCTATCACTGC |
| Atactin-2 | TGAGGGATGGCAACACTTTCCC |
| AtFT-1 | ACCCTCACCTCCGAGAATATCTCCAT |
| AtFT-2 | CTAAAGTCTTCTTCCTCCGCAGCCAC |
| AtSOC-1 | AGGCATACTAAGGATCGAGTCAGCAC |
| AtSOC-2 | GAAGAACAAGGTAACCCAATGAACAA |
| p3302-f | TGACGCACAATCCCACTATCCTT |
| p3302-r | CCGTCCAGCTCGACCAGGAT |
Table 1 Sequences of primers
| 名称 Primer name | 序列 Primer sequence (5′‒3′) |
|---|---|
| RAV1-f | ATGGATGGTAGTTACATAGATGAGA |
| RAV1-r | TTACAGAGCCCCAATTATCCTTTGT |
| 3302-R1-1 | GAACACGGGGGACTCTTGACATGGATGGTAGTTACATAGA |
| 3302-R1-2 | TTTACCCTCAGATCTACCATCAGAGCCCCAATTATCCTTT |
| RAV1-RT-1 | TAAAGGTTGGAGCCGATTTGTGA |
| RAV1-RT-2 | ACCCGTTTCTCGTATTCCAGTCA |
| CjGAPDH-1 | GGGAATCCTTGGTTACACTGAG |
| CjGAPDH-2 | ACCCCATTCGTTGTCATACC |
| Atactin-1 | ACCACTGTCCACTCTATCACTGC |
| Atactin-2 | TGAGGGATGGCAACACTTTCCC |
| AtFT-1 | ACCCTCACCTCCGAGAATATCTCCAT |
| AtFT-2 | CTAAAGTCTTCTTCCTCCGCAGCCAC |
| AtSOC-1 | AGGCATACTAAGGATCGAGTCAGCAC |
| AtSOC-2 | GAAGAACAAGGTAACCCAATGAACAA |
| p3302-f | TGACGCACAATCCCACTATCCTT |
| p3302-r | CCGTCCAGCTCGACCAGGAT |

Fig. 1 Phylogenetic tree and homology analysis of RAVs proteins from different speciesA: CDS sequence and encoded protein of CjRAV1. B: A phylogenetic tree of 15 RAVs proteins from different species (Cs: Camellia sinensis. Cl: Camellia lanceoleosa. Cj: Camellia japonica. At: Arabidopsis thaliana; Os: Oryza sativa). C: The sequence homology matrix of RAVs proteins in the indicative species

Fig. 5 Positive identification of CjRAV1 transgenic Arabidopsis linesA: PCR identification of DNA for positive strains of CjRAV-overexpressed Arabidopsis. B: Relative expressions of CjRAV1 intransgenic Arabidopsis detected by RT-qPCR. M: Marker. -: Negative control. +: Positive control. 1-8: From p3302-RAV1-1 to p3302-RAV1-8 transgenic Arabidopsis lines

Fig. 6 Comparative analysis of flowering time between wild type and CjRAV1 transgenic Arabidopsis plantsA: Phenotypic comparison between wild-type and CjRAV1 transgenic Arabidopsis plants. B: Flowering time statistics of wild-type and CjRAV1 transgenic Arabidopsis plants. C: Expressions of flowering regulatory genes in overexpressing CjRAV1 Arabidopsis. D: Total GAs content of wild type and CjRAV1 transgenic Arabidopsis plants. WT: Wild-type Arabidopsis; 1: p3302-RAV1-1 transgenic Arabidopsis line; 3: p3302-RAV1-3 transgenic Arabidopsis line
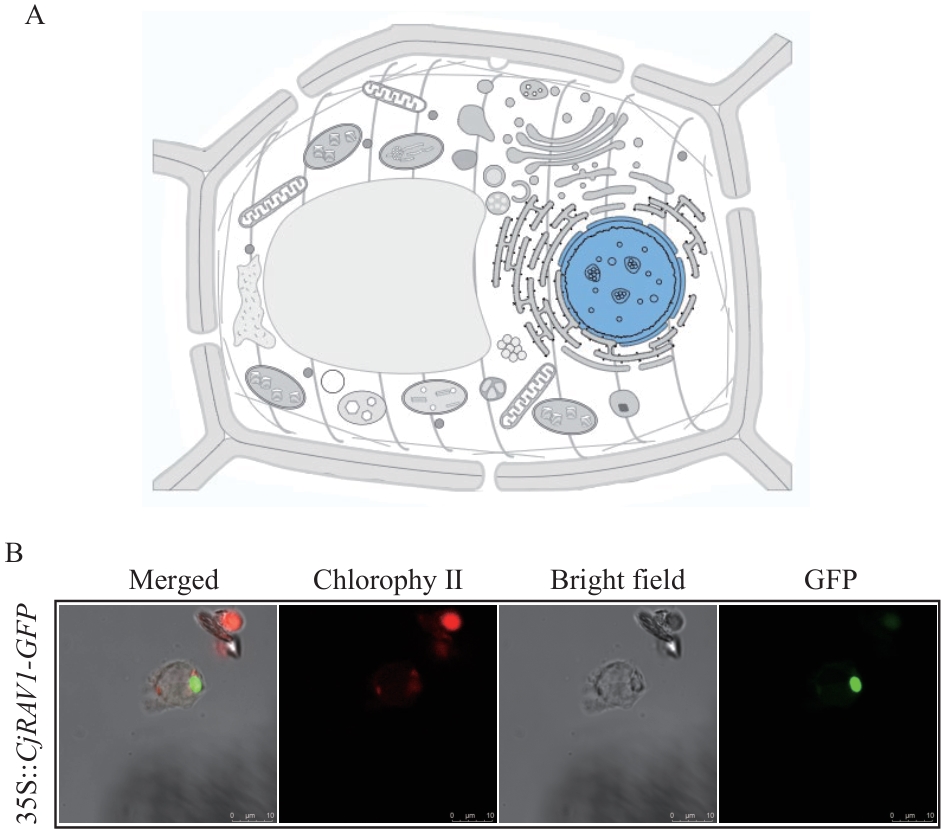
Fig. 7 Subcellular localization of the CjRAV1A: The localization prediction results of CjRAV1 in UniProt. B: Subcellular localization analysis of 35S::CjRAV1-GFP

Fig. 8 DAP-seq analysis of CjRAV1in C. japonicaA: Common peak Venn plot between repeated samples within the group. B: Distribution map of peak distance relative to gene TSS position. C: The largest DNA-motif bound by CjRAV1 in C. japonica. D: Distribution ratio diagram of peak on gene functional elements. E: KEGG enrichment pathway of the genes corresponding to all sites
结合序列 Motif | 结合峰 Peak name | 配对序列 Matched sequence | 得分 Score | 富集倍数 Fold enrichment |
|---|---|---|---|---|
| RMAGCAACAGM | CjRAV1_peak_6710 | GCAGCAACAAC | 14.956 5 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | CCAGCAGCAGA | 14.318 8 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | AAAGCAGCAGC | 14.217 4 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | CCATCAACAGA | 11.826 1 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | AAAGCAGCAAC | 11.391 3 | 31.405 34 |
Table 2 Potential downstream regulatory gene CjERF binding site of CjRAV1
结合序列 Motif | 结合峰 Peak name | 配对序列 Matched sequence | 得分 Score | 富集倍数 Fold enrichment |
|---|---|---|---|---|
| RMAGCAACAGM | CjRAV1_peak_6710 | GCAGCAACAAC | 14.956 5 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | CCAGCAGCAGA | 14.318 8 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | AAAGCAGCAGC | 14.217 4 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | CCATCAACAGA | 11.826 1 | 31.405 34 |
| RMAGCAACAGM | CjRAV1_peak_6710 | AAAGCAGCAAC | 11.391 3 | 31.405 34 |
| 1 | 徐剑, 聂瑞敏, 耿芳, 等. 观赏山茶属植物育种现状与展望 [J/OL]. 分子植物育种, 2024. . |
| Xu J, Nie RM, Geng F, et al. Current achievements and prospects of ornamental Camellia plants breeding [J/OL]. Mol Plant Breed, 2024. . | |
| 2 | 游慕贤. 中国古茶树 [J]. 国土绿化, 2002(2): 28. |
| You MX. China ancient tea tree [J]. Land Green, 2002(2): 28. | |
| 3 | 周利, 李玲莉, 宋春艳, 等. 重庆地区川山茶古树资源调查及保护现状分析[C]//中国环境科学学会, 中国植物学会. 2021年中国植物园学术年会论文集. 北京: 中国林业出版社, 2021: 80-84. |
| Zhou L, Li LL, Song CY, et al. Investigation and protection of ancient Camellia tree resources in Chongqing area [C]// Chinese Society for Environmental Sciences, Botanical Society of China. Proceedings of the 2021 Annual Conference of Chinese Botanical Gardens. Beijing: China Forestry Publishing House, 2021: 80-84. | |
| 4 | Blümel M, Dally N, Jung C. Flowering time regulation in crops-what did we learn from Arabidopsis? [J]. Curr Opin Biotechnol, 2015, 32: 121-129. |
| 5 | Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues [J]. Nat Rev Genet, 2012, 13(9): 627-639. |
| 6 | Boyle PE, Wisdom MM, Richardson MD. Testing flowering perennial plants in a bermudagrass (Cynodon spp.) lawn [J]. HortScience, 2020, 55(10): 1642-1646. |
| 7 | Hyun Y, Vincent C, Tilmes V, et al. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants [J]. Science, 2019, 363(6425): 409-412. |
| 8 | De La Torre AR, Piot A, Liu BB, et al. Functional and morphological evolution in gymnosperms: a portrait of implicated gene families [J]. Evol Appl, 2019, 13(1): 210-227. |
| 9 | Fletcher J, O’Connor-Moneley J, Frawley D, et al. Deletion of the Candida albicans TLO gene family using CRISPR-Cas9 mutagenesis allows characterisation of functional differences in α-, β- and γ- TLO gene function [J]. PLoS Genet, 2023, 19(12): e1011082. |
| 10 | Gu HJ, Pan ZJ, Jia MX, et al. Genome-wide identification and analysis of the cotton ALDH gene family [J]. BMC Genomics, 2024, 25(1): 513. |
| 11 | Osnato M, Castillejo C, Matías-Hernández L, et al. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis [J]. Nat Commun, 2012, 3: 808. |
| 12 | Matías-Hernández L, Aguilar-Jaramillo AE, Marín-González E, et al. RAV genes: regulation of floral induction and beyond [J]. Ann Bot, 2014, 114(7): 1459-1470. |
| 13 | Wang SM, Guo T, Shen YX, et al. Overexpression of MtRAV3 enhances osmotic and salt tolerance and inhibits growth of Medicago truncatula [J]. Plant Physiol Biochem, 2021, 163: 154-165. |
| 14 | Zhao L, Yang X, Du HL, et al. Reduced GA biosynthesis in GmRAV-transgenic tobacco causes the dwarf phenotype [J]. Russ J Plant Physiol, 2016, 63(5): 690-694. |
| 15 | Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis [J]. Nat Protoc, 2007, 2(7): 1565-1572. |
| 16 | Bartlett A, O'Malley RC, Huang SC, et al. Mapping genome-wide transcription-factor binding sites using DAP-seq [J]. Nat Protoc, 2017, 12(8): 1659-1672. |
| 17 | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method [J]. Nat Protoc, 2008, 3(6): 1101-1108. |
| 18 | Shin HY, Nam KH. RAV1 negatively regulates seed development by directly repressing MINI3 and IKU2 in Arabidopsis [J]. Mol Cells, 2018, 41(12): 1072-1080. |
| 19 | Osnato M, Matias-Hernandez L, Aguilar-Jaramillo AE, et al. Genes of the RAV family control heading date and carpel development in rice [J]. Plant Physiol, 2020, 183(4): 1663-1680. |
| 20 | Wang SM, Guo T, Wang Z, et al. Expression of three Related to ABI3/VP1 genes in Medicago truncatula caused increased stress resistance and branch increase in Arabidopsis thaliana [J]. Front Plant Sci, 2020, 11: 611. |
| 21 | Yuan YC, Zhou NN, Bai SS, et al. Evolutionary and integrative analysis of the gibberellin 20-oxidase, 3-oxidase, and 2-oxidase gene family in Paeonia ostii: insight into their roles in flower senescence [J]. Agronomy, 2024, 14(3): 590. |
| 22 | Xue YG, Zhang YT, Shan JM, et al. Growth repressor GmRAV binds to the GmGA3ox promoter to negatively regulate plant height development in soybean [J]. Int J Mol Sci, 2022, 23(3): 1721. |
| 23 | Li JL, Song CY, Li HM, et al. Comprehensive analysis of cucumber RAV family genes and functional characterization of CsRAV1 in salt and ABA tolerance in cucumber [J]. Front Plant Sci, 2023, 14: 1115874. |
| 24 | Hu HM, Tian S, Xie GH, et al. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis [J]. Proc Natl Acad Sci USA, 2021, 118(35): e2103895118. |
| 25 | Tao Z, Shen LS, Liu C, et al. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis [J]. Plant J, 2012, 70(4): 549-561. |
| 26 | Peng Z, Wang M, Zhang L, et al. EjRAV1/2 delay flowering through transcriptional repression of EjFTs and EjSOC1s in loquat [J]. Front Plant Sci, 2021, 12: 816086. |
| 27 | Chandler JW, Werr W. A phylogenetically conserved APETALA2/ETHYLENE RESPONSE FACTOR, ERF12, regulates Arabidopsis floral development [J]. Plant Mol Biol, 2020, 102(1/2): 39-54. |
| 28 | Huang YY, Xing XJ, Jin JY, et al. CmbHLH110, a novel bHLH transcription factor, accelerates flowering in Chrysanthemum [J]. Hortic Plant J, 2024, 10(6): 1437-1448. |
| 29 | Huang JG, Yang M, Liu P, et al. GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signalling in transgenic Arabidopsis [J]. Plant Cell Environ, 2009, 32(8): 1132-1145. |
| 30 | Yamasaki Y, Randall SK. Functionality of soybean CBF/DREB1 transcription factors [J]. Plant Sci, 2016, 246: 80-90. |
| 31 | Magome H, Yamaguchi S, Hanada A, et al. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis [J]. Plant J, 2008, 56(4): 613-626. |
| [1] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [2] | LIU Hong-li, MA Yi-dan, WANG Wan-ru, YANG Ya-ru, HE Dan, LIU Yi-ping, KONG De-zheng. Cloning and Functional Analysis of NnCYP707A1 Gene from Lotus [J]. Biotechnology Bulletin, 2025, 41(5): 197-207. |
| [3] | TIAN Qin, LIU Kui, WU Xiang-wei, JI Yuan-yuan, CAO Yi-bo, ZHANG Ling-yun. Functional Study of Transcription Factor VcMYB17 in Regulating Drought Tolerance in Blueberry [J]. Biotechnology Bulletin, 2025, 41(4): 198-210. |
| [4] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [5] | CHEN Ying-e, LIANG Qiao-lan. Research Progress in the Effects of Plant Abscisic Acid and Its Receptor Gene PYL9 [J]. Biotechnology Bulletin, 2024, 40(12): 1-11. |
| [6] | HOU Ying-xiang, FEI Si-tian, SONG Song-quan, LUO Yong, ZHANG Chao. Research Progress in MADS-box Family in Rice [J]. Biotechnology Bulletin, 2024, 40(11): 103-112. |
| [7] | WANG Gui-fang, YAO Yuan-tao, XU Hai-feng, XIANG Kun, LIANG Jia-hui, ZHANG Shu-hui, WANG Wen-ru, ZHANG Ming-juan, ZHANG Mei-yong, CHEN Xin. The Gene JrSnRK1α1.1 of Walnut Regulates Seed Oil Synthesis and Accumulation [J]. Biotechnology Bulletin, 2023, 39(9): 183-191. |
| [8] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [9] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [10] | ZHANG Hao, LI Zhe, GUO Kai, HUANG Yan-hua, HAO Yong-ren. Functional Analysis of TvGCN5 Gene Encoding Histone Acetylase from Trichoderma viride Tv-1511 [J]. Biotechnology Bulletin, 2022, 38(5): 136-148. |
| [11] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [12] | LIU Sha-yu, CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu. Biological Function of a Zn2Cys6 Transcription Factor CgAswA in Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2021, 37(9): 161-170. |
| [13] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [14] | YIN Chao-min, FAN Xiu-zhi, SHI De-fang, GAO Hong. CRISPR/Cas Genome Editing Technology and Its Application in Fungi [J]. Biotechnology Bulletin, 2017, 33(3): 58-65. |
| [15] | Su Zijing, Li Qiaoling, Huang Cheng, Xie Chengjian, Yang Xingyong. RNAi Technology and Its Application in Fungal Gene Functional Studies [J]. Biotechnology Bulletin, 2015, 31(8): 50-58. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||