








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 106-116.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1077
Previous Articles Next Articles
HAN Yi1( ), HOU Chang-lin1, TANG Lu1, SUN Lu1, XIE Xiao-dong1, LIANG Chen2(
), HOU Chang-lin1, TANG Lu1, SUN Lu1, XIE Xiao-dong1, LIANG Chen2( ), CHEN Xiao-qiang1(
), CHEN Xiao-qiang1( )
)
Received:2024-11-06
Online:2025-07-26
Published:2025-07-22
Contact:
LIANG Chen, CHEN Xiao-qiang
E-mail:gym604666@163.com;2008lc22@163.com;chenxia9663@126.com
HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare[J]. Biotechnology Bulletin, 2025, 41(7): 106-116.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | Tm (℃) | 用途 Purpose |
|---|---|---|---|
| HvERECTA-F | ACGGTGACTCTCCGCAGC | 58.8 | 基因扩增 Gene amplification |
| HvERECTA-R | CGACTACCCGAATCATCCTCC | 60.9 | |
| qHvERECTA-F | TGGCATTGCTAAGAGTTTGTGT | 57.9 | RT-qPCR |
| qHvERECTA-R | GATTGCACTCGTTGTCCACG | 59.4 | |
| qHvSPCH-F | CCGAAGACTTATTCAGCATCC | 56.7 | |
| qHvSPCH-R | CTCATGCGATATTCCGTCAC | 54.7 | |
| qHvMUTE-F | TCCTTCGTCCTCAAGATTGG | 57.4 | |
| qHvMUTE-R | ATATCGCCATGGATGAGTAGTC | 56.1 | |
| qFAMA-F | TGGCGGAGAACAAGTCGAG | 59.2 | |
| qFAMA-R | GTGTGGAGGATGGACATGTG | 55.8 | |
| Actin-F | TGGATCGGAGGGTCCATCCT | 63.5 |
Table 1 Primers used in this study and their uses
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | Tm (℃) | 用途 Purpose |
|---|---|---|---|
| HvERECTA-F | ACGGTGACTCTCCGCAGC | 58.8 | 基因扩增 Gene amplification |
| HvERECTA-R | CGACTACCCGAATCATCCTCC | 60.9 | |
| qHvERECTA-F | TGGCATTGCTAAGAGTTTGTGT | 57.9 | RT-qPCR |
| qHvERECTA-R | GATTGCACTCGTTGTCCACG | 59.4 | |
| qHvSPCH-F | CCGAAGACTTATTCAGCATCC | 56.7 | |
| qHvSPCH-R | CTCATGCGATATTCCGTCAC | 54.7 | |
| qHvMUTE-F | TCCTTCGTCCTCAAGATTGG | 57.4 | |
| qHvMUTE-R | ATATCGCCATGGATGAGTAGTC | 56.1 | |
| qFAMA-F | TGGCGGAGAACAAGTCGAG | 59.2 | |
| qFAMA-R | GTGTGGAGGATGGACATGTG | 55.8 | |
| Actin-F | TGGATCGGAGGGTCCATCCT | 63.5 |

Fig. 3 Conservative structure analysis of HvERECTAA: Multiple sequence alignment (STK_BAK1_like by black dashed line, ATP binding site by red box. Hv: Hordeum vulgare; Ta: Triticum aestivum; Bd: Brachypodium distachyon; Sb: Sorghum bicolo; Zm: Zea mays; Qs: Oryza sativa Japonica Group; Gm: Glycine max; At: Arabidopsis thaliana). B: Analysis of conservative domain. C: Prediction of conservative base sequence

Fig. 5 Prediction of protein interactions of HvERECTALine colors indicate the type of interactive evidence and the number of lines indicates the strength of data support

Fig. 6 Phylogenetic tree of HvERECTA and other homologous gene ERfsThe blue sector is ERECTA, the red sector is ERL, the red branch is grasses, and the blue branch is dicotyledonous
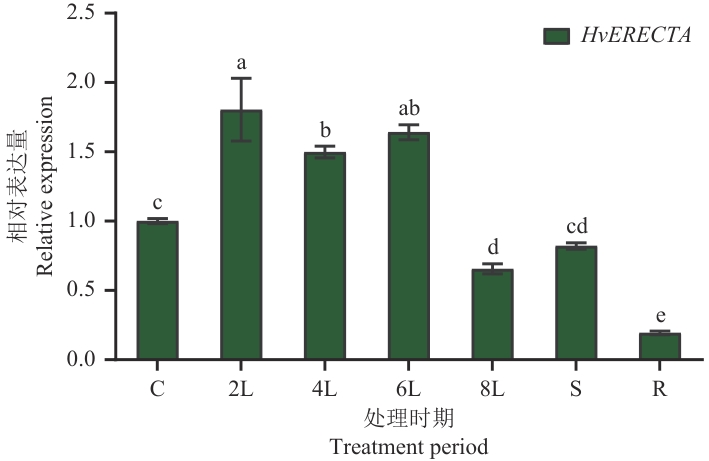
Fig. 8 Relative expressions of HvERECTA in different organs of barleyC: Coleoptile; 2L: 2 d leaf age; 4L: 4 d leaf age; 6L: 6 d leaf age; 8L: 8 d leaf age; S: stem; R: root. Lowercase letters for significant differences (P<0.05)
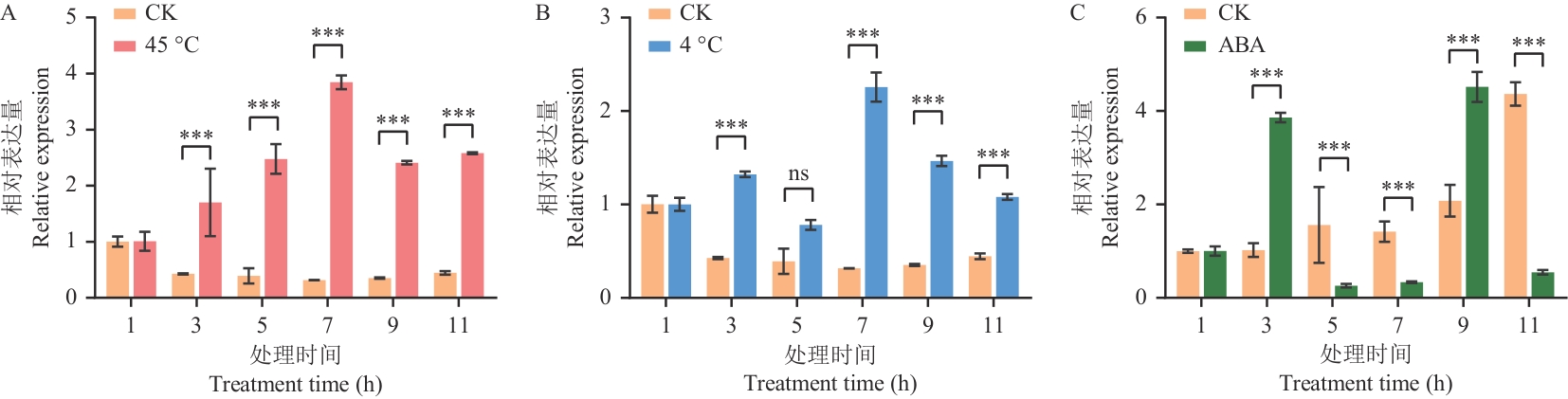
Fig. 9 Expressions of HvERECTA gene in barley treated with ABA and under high temperature, low temperatureA: High temperature treatment. B: Low temperature treatment. C: ABA treatment.* indicates significant difference in gene expression between treatment group and control group (0.01≤P<0.05), ** indicating that the difference in gene expression between the treatment group and the control group was significant (0.001≤P<0.01), *** indicating that the difference in gene expression between the treatment and control groups was extremely significant (P<0.001).nsindicates no significant difference in gene expression between treatment group and control group (P>0.05). The same below
| [1] | Shirvani H, Mehrabi AA, Farshadfar M, et al. Investigation of the morphological, physiological, biochemical, and catabolic characteristics and gene expression under drought stress in tolerant and sensitive genotypes of wild barley [Hordeum vulgare subsp. spontaneum (K. Koch) Asch. & Graebn. [J]. BMC Plant Biol, 2024, 24(1): 214. |
| [2] | 赵盟, 王春超, 张仁旭, 等. 中国大麦育成品种产量相关性状鉴定评价 [J]. 植物遗传资源学报, 2022, 23(5): 1371-1382. |
| Zhao M, Wang CC, Zhang RX, et al. Evaluation of the yield-related traits of Chinese barley cultivars [J]. J Plant Genet Resour, 2022, 23(5): 1371-1382. | |
| [3] | 阿丽亚古丽·艾合买提. 玉米ZmNAC3基因的克隆及生物学功能的初步研究 [D]. 曲阜: 曲阜师范大学, 2024. |
| Aliaguri A. Cloning and preliminary study on biological function of maize ZmNAC3 gene [D]. Qufu: Qufu Normal University, 2024. | |
| [4] | Zhang CJ, Ren YY, Cao LJ, et al. Characteristics of dry-wet climate change in China during the past 60 years and its trends projection [J]. Atmosphere, 2022, 13(2): 275. |
| [5] | Doll Y, Koga H, Tsukaya H. Experimental validation of the mechanism of stomatal development diversification [J]. J Exp Bot, 2023, 74(18): 5667-5681. |
| [6] | Berchembrock YV, Botelho FBS, Srivastava V. Suppression of ERECTA signaling impacts agronomic performance of soybean (Glycine max (L) merril) in the greenhouse [J]. Front Plant Sci, 2021, 12: 667825. |
| [7] | Masle J, Gilmore SR, Farquhar GD. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis [J]. Nature, 2005, 436(7052): 866-870. |
| [8] | 张玉涵. 水稻ERECTA基因家族的进化分析及功能鉴定 [D]. 南京: 南京大学, 2018. |
| Zhang YH. Evolutionary analysis and functional identification of rice ERECTA gene family [D]. Nanjing: Nanjing University, 2018. | |
| [9] | 赵雪雅, 张坤鹏, 张汇东, 等. ERECTA基因家族调控植物形态发育机制研究进展 [J]. 沈阳农业大学学报, 2023, 54(2): 231-238. |
| Zhao XY, Zhang KP, Zhang HD, et al. Research progress on the mechanism of ERECTA gene family regulating plant morphological development [J]. J Shenyang Agric Univ, 2023, 54(2): 231-238. | |
| [10] | Shpak ED, Lakeman MB, Torii KU. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape [J]. Plant Cell, 2003, 15(5): 1095-1110. |
| [11] | Shpak ED, Berthiaume CT, Hill EJ, et al. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation [J]. Development, 2004, 131(7): 1491-1501. |
| [12] | Lee JS, Kuroha T, Hnilova M, et al. Direct interaction of ligand-receptor pairs specifying stomatal patterning [J]. Genes Dev, 2012, 26(2): 126-136. |
| [13] | 陈春林, 李白雪, 李金玲, 等. 甜瓜CmEPF基因家族的鉴定及表达分析 [J]. 生物技术通报, 2024, 40(4): 130-138. |
| Chen CL, Li BX, Li JL, et al. Identification and expression analysis of epidermal patterning factor (EPF) genes in Cucumis melo [J]. Biotechnol Bull, 2024, 40(4): 130-138. | |
| [14] | Jewaria PK, Hara T, Tanaka H, et al. Differential effects of the peptides Stomagen, EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level [J]. Plant Cell Physiol, 2013, 54(8): 1253-1262. |
| [15] | MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage [J]. Nature, 2007, 445(7127): 537-540. |
| [16] | Qi XY, Han SK, Dang JH, et al. Autocrine regulation of stomatal differentiation potential by EPF1 and ERECTA-LIKE1 ligand-receptor signaling [J]. eLife, 2017, 6: e24102. |
| [17] | Pillitteri LJ, Bemis SM, Shpak ED, et al. Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development [J]. Development, 2007, 134(17): 3099-3109. |
| [18] | 刘梦龙, 魏健, 任姿蓉, 等. 气孔研究进展: 从保卫细胞到光合作用 [J]. 生命科学, 2024, 36(10): 1289-1304. |
| Liu ML, Wei J, Ren ZR, et al. Research progress of stomata: from guard cells to photosynthesis [J]. Chin Bull Life Sci, 2024, 36(10): 1289-1304. | |
| [19] | 侯笑颜, 孙美红, 张恒, 等. 植物ERECTA基因家族特征及其非生物胁迫功能 [J]. 上海师范大学学报: 自然科学版, 2020, 49(6): 719-730. |
| Hou XY, Sun MH, Zhang H, et al. Research advances of ERECTA gene family and its functions in abiotic stress response in plants [J]. J Shanghai Norm Univ Nat Sci, 2020, 49(6): 719-730. | |
| [20] | Li HG, Yang YL, Wang HL, et al. The receptor-like kinase ERECTA confers improved water use efficiency and drought tolerance to poplar via modulating stomatal density [J]. Int J Mol Sci, 2021, 22(14): 7245. |
| [21] | Juneidi S, Gao ZY, Yin HR, et al. Breaking the summer dormancy of Pinellia ternata by introducing a heat tolerance receptor-like kinase ERECTA gene [J]. Front Plant Sci, 2020, 11: 780. |
| [22] | 次尔甲玛, 王宏鹏, 苟筱颖, 等. 大麦气孔发育相关基因HvSPCH的克隆及初步功能分析 [J]. 麦类作物学报, 2022, 42(10): 1175-1181. |
| Ci E, Wang HP, Gou XY, et al. Cloning and functional analysis of stomatal development related gene HvSPCH in barley [J]. J Triticeae Crops, 2022, 42(10): 1175-1181. | |
| [23] | 王宏鹏, 曹高燚, 李明, 等. 大麦气孔发育转录因子基因HvMUTE的克隆及生物学功能初步鉴定 [J]. 华北农学报, 2023, 38(1): 40-45. |
| Wang HP, Cao GY, Li M, et al. Gene cloning and preliminary function identification of HvMUTE transcription factor for barley stomatal development [J]. Acta Agric Boreali Sin, 2023, 38(1): 40-45. | |
| [24] | 孙璐, 蔡浩宇, 李明, 等. 大麦气孔发育相关基因HvFAMA的克隆及功能初步分析 [J]. 麦类作物学报, 2024, 44(11): 1397-1403. |
| Sun L, Cai HY, Li M, et al. Cloning and preliminary functional analysis of HvFAMA gene related to stomatal development in Hordeum vulgare [J]. J Triticeae Crops, 2024, 44(11): 1397-1403. | |
| [25] | Gou XP, He K, Yang H, et al. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana [J]. BMC Genomics, 2010, 11: 19. |
| [26] | Kosentka PZ, Zhang L, Simon YA, et al. Identification of critical functional residues of receptor-like kinase ERECTA [J]. J Exp Bot, 2017, 68(7): 1507-1518. |
| [27] | Gish LA, Clark SE. The RLK/Pelle family of kinases [J]. Plant J, 2011, 66(1): 117-127. |
| [28] | Chen LL, Cochran AM, Waite JM, et al. Direct attenuation of Arabidopsis ERECTA signalling by a pair of U-box E3 ligases [J]. Nat Plants, 2023, 9(1): 112-127. |
| [29] | 刘敏. 葡萄幼苗高温响应机理的研究及ERECTA基因家族的表达和功能分析 [D]. 杨凌: 西北农林科技大学, 2018. |
| Liu M. Study on the mechanism of high temperature response of grape seedlings and the expression and function analysis of ERECTA gene family [D]. Yangling: Northwest A & F University, 2018. | |
| [30] | 李昕雨, 王泽博, 邱颖胜, 等. 植物启动子响应非生物逆境胁迫研究进展 [J]. 种子科技, 2022, 40(3): 16-18. |
| Li XY, Wang ZB, Qiu YS, et al. Research progress of plant promoters responding to abiotic stress [J]. Seed Sci Technol, 2022, 40(3): 16-18. | |
| [31] | Koua AP, Oyiga BC, Baig MM, et al. Breeding driven enrichment of genetic variation for key yield components and grain starch content under drought stress in winter wheat [J]. Front Plant Sci, 2021, 12: 684205. |
| [32] | Zhang MM, Zhang SQ. Stomatal development: NRPM proteins in dynamic localization of ERECTA receptor [J]. Curr Biol, 2024, 34(4): R143-R146. |
| [33] | 刘敏, 房玉林. 葡萄ERECTA基因过表达提高拟南芥生长量和耐热性 [J]. 生物技术通报, 2020, 36(6): 46-53. |
| Liu M, Fang YL. Overexpression of grape ERECTA enhances growth and thermo-tolerance of Arabidopsis [J]. Biotechnol Bull, 2020, 36(6): 46-53. | |
| [34] | Hughes J, Hepworth C, Dutton C, et al. Reducing stomatal density in barley improves drought tolerance without impacting on yield [J]. Plant Physiol, 2017, 174(2): 776-787. |
| [35] | Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development [J]. Plant Cell, 2006, 18(10): 2493-2505. |
| [36] | McKown KH, Ximena Anleu Gil M, Mair A, et al. Expanded roles and divergent regulation of FAMA in Brachypodium and Arabidopsis stomatal development [J]. Plant Cell, 2023, 35(2): 756-775. |
| [37] | Ortega A, de Marcos A, Illescas-Miranda J, et al. The tomato genome encodes SPCH, MUTE, and FAMA candidates that can replace the endogenous functions of their Arabidopsis orthologs [J]. Front Plant Sci, 2019, 10: 1300. |
| [1] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| [2] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [3] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [4] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [5] | WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family [J]. Biotechnology Bulletin, 2025, 41(6): 87-98. |
| [6] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [7] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [8] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [9] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [10] | HAN Jiang-tao, ZHANG Shuai-bo, QIN Ya-rui, HAN Shuo-yang, ZHANG Ya-kang, WANG Ji-qing, DU Qing-jie, XIAO Huai-juan, LI Meng. Identification of β-amylase Gene Family in Melon and Their Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(3): 171-180. |
| [11] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [12] | GE Shi-jie, LIU Yi-de, ZHANG Hua-dong, NING Qiang, ZHU Zhan-wang, WANG Shu-ping, LIU Yi-ke. Identification and Expression Analysis of Protein Disulfide Isomerase Gene Family in Wheat [J]. Biotechnology Bulletin, 2025, 41(2): 85-96. |
| [13] | YIN Yuan, CHENG Shuang, LIU Ding-hao, DENG Xiao-xia, LI Kai-yue, WANG Jing-hong, LIN Ji-xiang. Research Progress in Exogenous Hydrogen Peroxide(H2O2)Affecting Plant Growth and Physiological Metabolism under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(1): 1-13. |
| [14] | WU Zhi-jian, LIU Guang-yang, LIN Zhi-hao, SHENG Bin, CHEN Ge, XU Xiao-min, WANG Jun-wei, XU Dong-hui. Research Progress of Nano-regulation of Vegetable Seed Germination and Its Mechanism [J]. Biotechnology Bulletin, 2025, 41(1): 14-24. |
| [15] | LI Yu-xin, LI Miao, DU Xiao-fen, HAN Kang-ni, LIAN Shi-chao, WANG Jun. Identification and Expression Analysis of SiSAP Gene Family in Foxtail Millet(Setaria italica) [J]. Biotechnology Bulletin, 2025, 41(1): 143-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||