








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 95-105.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0109
Previous Articles Next Articles
NIU Jing-ping1( ), ZHAO Jing2, GUO Qian2, WANG Shu-hong2, ZHAO Jin-zhong3, DU Wei-jun2, YIN Cong-cong3, YUE Ai-qin2(
), ZHAO Jing2, GUO Qian2, WANG Shu-hong2, ZHAO Jin-zhong3, DU Wei-jun2, YIN Cong-cong3, YUE Ai-qin2( )
)
Received:2025-01-26
Online:2025-07-26
Published:2025-07-22
Contact:
YUE Ai-qin
E-mail:niujingping@sxau.edu.cn;yueaiqinnd@126.com
NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA[J]. Biotechnology Bulletin, 2025, 41(7): 95-105.
基因名称 Gene name | 基因ID Gene ID | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|---|
| GmNAC030 | Glyma.05G195000 | GGTTCAGTTCCAGCACGAGA | CGTGTCCATGTACAATTGGTCG |
| GmNAC043 | Glyma.06G248900 | TCTTCCCGCAAACACAACCT | TGACCCATTGCTGCATTCCA |
| GmNAC085 | Glyma.12G149100 | CGACATGCTGGAATCGTTGC | GTCCTGCTGCTGCATTGTTC |
| GmNAC092 | Glyma.12G221500 | ACAACTCGACGACGTTCTGG | CCCCCTTCACCCAAGTTCTG |
| GmNAC101 | Glyma.13G279900 | AACACGCTGCAACAACAACA | ATTCCCCGAAGCGAAATCGG |
| tublin | GGAGTTCACAGAGGCAGAG | CACTTACGCATCACATAGC |
Table 1 Information of primers for RT-qPCR
基因名称 Gene name | 基因ID Gene ID | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|---|
| GmNAC030 | Glyma.05G195000 | GGTTCAGTTCCAGCACGAGA | CGTGTCCATGTACAATTGGTCG |
| GmNAC043 | Glyma.06G248900 | TCTTCCCGCAAACACAACCT | TGACCCATTGCTGCATTCCA |
| GmNAC085 | Glyma.12G149100 | CGACATGCTGGAATCGTTGC | GTCCTGCTGCTGCATTGTTC |
| GmNAC092 | Glyma.12G221500 | ACAACTCGACGACGTTCTGG | CCCCCTTCACCCAAGTTCTG |
| GmNAC101 | Glyma.13G279900 | AACACGCTGCAACAACAACA | ATTCCCCGAAGCGAAATCGG |
| tublin | GGAGTTCACAGAGGCAGAG | CACTTACGCATCACATAGC |
转录因子家族 TF family | 基因数 Gene number | 转录因子家族 TF family | 基因数 Gene number |
|---|---|---|---|
| MYB | 335 | MIKC | 84 |
| ERF | 305 | Dof | 79 |
| bHLH | 295 | C3H | 69 |
| MYB_related | 266 | GATA | 60 |
| HB-other | 204 | ARF | 55 |
| WRKY | 176 | TCP | 54 |
| NAC | 171 | C2H2 | 53 |
| M_type | 144 | HSF | 52 |
| bZIP | 144 | FAR1 | 49 |
| B3 | 131 | ZF-HD | 47 |
| GRAS | 111 | SBP | 45 |
| LBD | 91 | AP2 | 44 |
| HD-ZIP | 41 | CPP | 12 |
| DBB | 34 | EIL | 11 |
| Nin-like | 28 | BBR-BPC | 10 |
| CO-like | 22 | GeBP | 9 |
| GRF | 22 | LSD | 8 |
| NF-YA | 21 | Whirly | 7 |
| SRS | 21 | HB-PHD | 6 |
| TALE | 18 | NF-X1 | 5 |
| YABBY | 17 | RAV | 4 |
| BES1 | 15 | S1Fa-like | 4 |
| CAMTA | 15 | LFY | 2 |
| E2F/DP | 14 | NZZ/SPL | 1 |
Table 2 Statistics of transcription factors (TF)
转录因子家族 TF family | 基因数 Gene number | 转录因子家族 TF family | 基因数 Gene number |
|---|---|---|---|
| MYB | 335 | MIKC | 84 |
| ERF | 305 | Dof | 79 |
| bHLH | 295 | C3H | 69 |
| MYB_related | 266 | GATA | 60 |
| HB-other | 204 | ARF | 55 |
| WRKY | 176 | TCP | 54 |
| NAC | 171 | C2H2 | 53 |
| M_type | 144 | HSF | 52 |
| bZIP | 144 | FAR1 | 49 |
| B3 | 131 | ZF-HD | 47 |
| GRAS | 111 | SBP | 45 |
| LBD | 91 | AP2 | 44 |
| HD-ZIP | 41 | CPP | 12 |
| DBB | 34 | EIL | 11 |
| Nin-like | 28 | BBR-BPC | 10 |
| CO-like | 22 | GeBP | 9 |
| GRF | 22 | LSD | 8 |
| NF-YA | 21 | Whirly | 7 |
| SRS | 21 | HB-PHD | 6 |
| TALE | 18 | NF-X1 | 5 |
| YABBY | 17 | RAV | 4 |
| BES1 | 15 | S1Fa-like | 4 |
| CAMTA | 15 | LFY | 2 |
| E2F/DP | 14 | NZZ/SPL | 1 |

Fig. 2 Clustering diagram of samplesA: Clustering diagram of all 36 samples. The three outlier samples are written in red fonts. B: Clustering diagram of 33 samples after removing the three outlier samples

Fig. 3 Determination of soft threshold power (β value) via WGCNA and identification of modulesA: Determination of soft threshold power (β value). B: Gene module classification, dendrogram indicate genes and different colors indicate different modules. C: Heat map of correlations between resistance trait and modules. The correlation coefficients are shown above each cell, and the P values are shown below

Fig. 5 Expression profiles of NAC and validation of NAC by RT-qPCRA: Expression profiles of five NAC at different time point X149 and X97. B: Validation of GmNAC030 in X149 by RT-qPCR; *: P<0.05, **: P<0.01,The same below
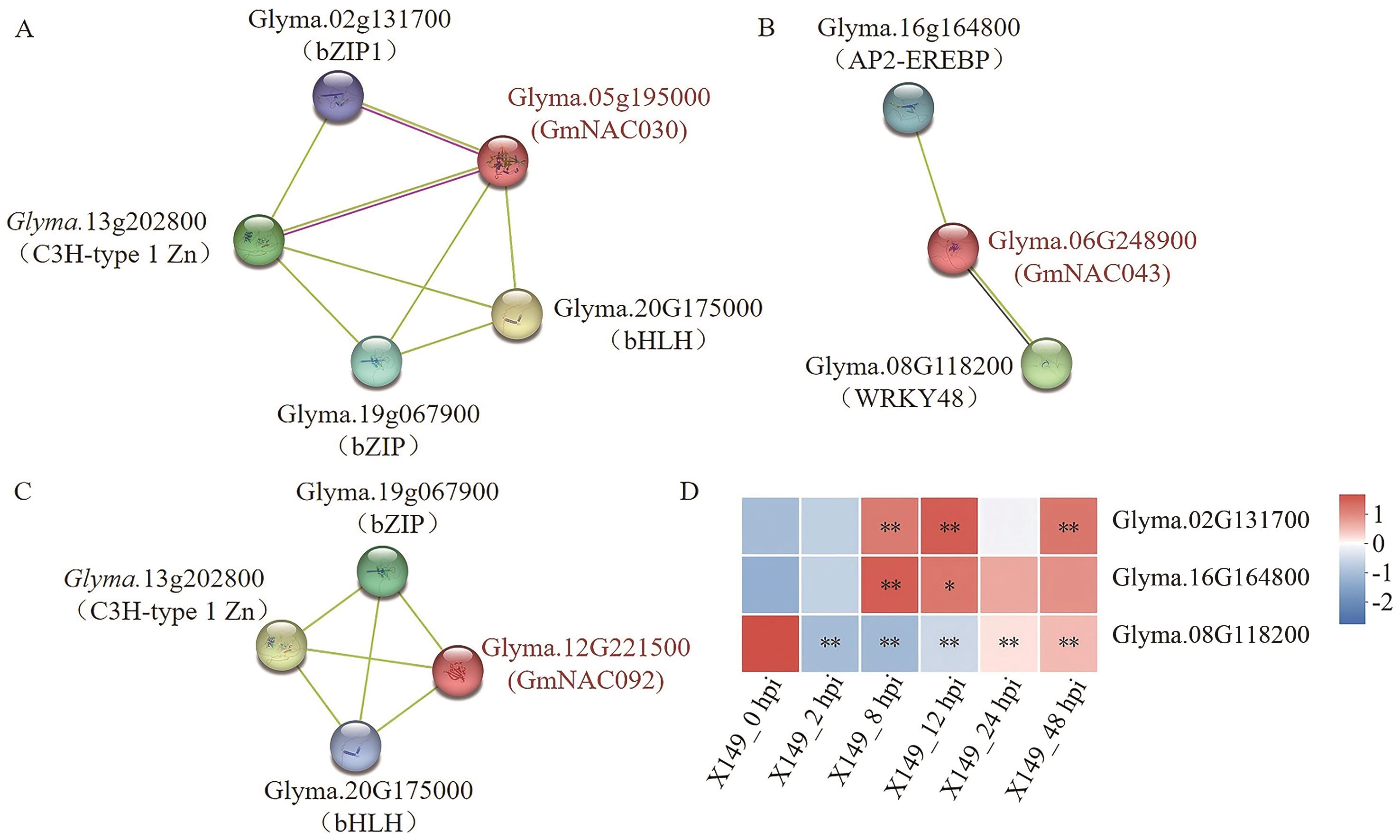
Fig. 6 Analysis of NAC protein interaction and expression profiles of interacting proteins in transcriptomeA: Interacting protein Glyma.05g195000. B: Interacting protein Glyma.06G248900. C: Interacting protein Glyma.12G221500. D: Expression profiles of interacting protein in the modules
| [1] | 王大刚, 李凯, 智海剑. 大豆抗大豆花叶病毒病基因研究进展 [J]. 中国农业科学, 2018, 51(16): 3040-3059. |
| Wang DG, Li K, Zhi HJ. Progresses of resistance on soybean mosaic virus in soybean [J]. Sci Agric Sin, 2018, 51(16): 3040-3059. | |
| [2] | 高乐, 李志强, 李凯, 等. 大豆花叶病毒病转基因抗性研究进展 [J]. 中国油料作物学报, 2022, 44(2): 434-441. |
| Gao L, Li ZQ, Li K, et al. Advances in transgenic resistance to soybean mosaic virus disease [J]. Chin J Oil Crop Sci, 2022, 44(2): 434-441. | |
| [3] | 刘惠玲, 江炳玉, 张静, 等. 植物NAC转录因子的研究进展 [J]. 福建农林大学学报: 自然科学版, 2024, 53(6): 742-753. |
| Liu HL, Jiang BY, Zhang J, et al. Advances in study on plant NAC transcription factors [J]. J Fujian Agric For Univ: Nat Sci Ed, 2024, 53(6): 742-753. | |
| [4] | Todeschini AL, Georges A, Veitia RA. Transcription factors: specific DNA binding and specific gene regulation [J]. Trends Genet, 2014, 30(6): 211-219. |
| [5] | Baillo EH, Kimotho RN, Zhang ZB, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement [J]. Genes, 2019, 10(10): 771. |
| [6] | Zhang CQ, Grosic S, Whitham SA, et al. The requirement of multiple defense genes in soybean Rsv1-mediated extreme resistance to soybean mosaic virus [J]. Mol Plant Microbe Interact, 2012, 25(10): 1307-1313. |
| [7] | 钟晨丽, 兰胡娇, 王文絮, 等. GmWRKY33A正向调控大豆抗病性 [J]. 生物工程学报, 2024, 40(10): 3810-3822. |
| Zhong CL, Lan HJ, Wang WX, et al. GmWRKY33A positively regulates disease resistance insoybean (Glycine max) [J]. Chin J | |
| Biotechnol, 2024, 40(10): 3810-3822. | |
| [8] | 钟晨丽, 王文絮, 廖莉娜, 等. 沉默大豆GmWRKY33B基因导致大豆抗病性降低 [J]. 生物工程学报, 2024, 40(1): 163-176. |
| Zhong CL, Wang WX, Liao LN, et al. Silencing GmWRKY33B genes leads to reduced disease resistance in soybean [J]. Chin J Biotechnol, 2024, 40(1): 163-176. | |
| [9] | Ren QY, Jiang H, Xiang WY, et al. A MADS-box gene is involved in soybean resistance to multiple Soybean mosaic virus strains [J]. Crop J, 2022, 10(3): 802-808. |
| [10] | Li H, Liu JY, Yuan XX, et al. Comparative transcriptome analysis reveals key pathways and regulatory networks in early resistance of Glycine max to soybean mosaic virus [J]. Front Microbiol, 2023, 14: 1241076. |
| [11] | DeMers LC, Redekar NR, Kachroo A, et al. A transcriptional regulatory network of Rsv3-mediated extreme resistance against Soybean mosaic virus [J]. PLoS One, 2020, 15(4): e0231658. |
| [12] | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis [J]. BMC Bioinformatics, 2008, 9: 559. |
| [13] | Zhu HH, Li RQ, Fang YY, et al. Weighted gene co-expression network analysis uncovers critical genes and pathways involved in soybean response to soybean mosaic virus [J]. Agronomy, 2024, 14(11): 2455. |
| [14] | Niu JP, Zhao J, Guo Q, et al. WGCNA reveals hub genes and key gene regulatory pathways of the response of soybean to infection by Soybean mosaic virus [J]. Genes, 2024, 15(5): 566. |
| [15] | Burke R, Schwarze J, Sherwood OL, et al. Stressed to death: the role of transcription factors in plant programmed cell death induced by abiotic and biotic stimuli [J]. Front Plant Sci, 2020, 11: 1235. |
| [16] | Masri R, Kiss E. The role of NAC genes in response to biotic stresses in plants [J]. Physiol Mol Plant Pathol, 2023, 126: 102034. |
| [17] | Viswanath KK, Kuo SY, Tu CW, et al. The role of plant transcription factors in the fight against plant viruses [J]. Int J Mol Sci, 2023, 24(9): 8433. |
| [18] | Wang X, Goregaoker SP, Culver JN. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses [J]. J Virol, 2009, 83(19): 9720-9730. |
| [19] | Xie Q, Sanz-Burgos AP, Guo H, et al. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein [J]. Plant Mol Biol, 1999, 39(4): 647-656. |
| [20] | Huang Y, Li T, Xu ZS, et al. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars [J]. Plant Physiol Biochem, 2017, 120: 61-74. |
| [21] | Selth LA, Dogra SC, Saif Rasheed M, et al. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication [J]. Plant Cell, 2005, 17(1): 311-325. |
| [22] | Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks [J]. Genome Res, 2003, 13(11): 2498-2504. |
| [23] | Rui R, Liu SC, Karthikeyan A, et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to soybean mosaic virus [J]. Theor Appl Genet, 2017, 130(11): 2395-2410. |
| [24] | Irsigler AST, Costa MDL, Zhang P, et al. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways [J]. BMC Genomics, 2007, 8: 431. |
| [25] | Melo BP, Fraga OT, Silva JCF, et al. Revisiting the soybean GmNAC superfamily [J]. Front Plant Sci, 2018, 9: 1864. |
| [26] | Mendes GC, Reis PAB, Calil IP, et al. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme [J]. Proc Natl Acad Sci USA, 2013, 110(48): 19627-19632. |
| [27] | Melo BP, Lourenço-Tessutti IT, Fraga OT, et al. Contrasting roles of GmNAC065 and GmNAC085 in natural senescence, plant development, multiple stresses and cell death responses [J]. Sci Rep, 2021, 11(1): 11178. |
| [28] | Liu WJ, Mei ZX, Yu L, et al. The ABA-induced NAC transcription factor MdNAC1 interacts with a bZIP-type transcription factor to promote anthocyanin synthesis in red-fleshed apples [J]. Hortic Res, 2023, 10(5): uhad049. |
| [29] | Yu D, Wei W, Fan ZQ, et al. VabHLH137 promotes proanthocyanidin and anthocyanin biosynthesis and enhances resistance to Colletotrichum gloeosporioides in grapevine [J]. Hortic Res, 2022, 10(2): uhac261. |
| [30] | Zhong Q, Yu JT, Wu YD, et al. Rice transcription factor OsNAC2 maintains the homeostasis of immune responses to bacterial blight [J]. Plant Physiol, 2024, 195(1): 785-798. |
| [31] | Wang H, Bi Y, Gao YZ, et al. A Pathogen-Inducible rice nac transcription factor ONAC096 contributes to immunity against Magnaprothe oryzae and Xanthomonas oryzae pv. oryzae by direct binding to the promoters of OsRap2.6, OsWRKY62, and OsPAL1 [J]. Front Plant Sci, 2021, 12: 802758. |
| [32] | Gao SQ, Chen M, Xu ZS, et al. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants [J]. Plant Mol Biol, 2011, 75(6): 537-553. |
| [33] | 田蕊, 张华, 黄玫红, 等. 大豆抗旱遗传位点及候选基因发掘 [J]. 中国农业科技导报, 2023, 25(9): 69-82. |
| Tian R, Zhang H, Huang MH, et al. Mining of candidate genes and genetic loci conferring drought tolerance in soybean [J]. J Agric Sci Technol, 2023, 25(9): 69-82. | |
| [34] | Singh V, Roy S, Singh D, et al. Arabidopsis flowering locus D influences systemic-acquired-resistance- induced expression and histone modifications of WRKY genes [J]. J Biosci, 2014, 39(1): 119-126. |
| [35] | Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants [J]. Front Microbiol, 2013, 4: 248. |
| [36] | Freitas EO, Melo BP, Lourenço-Tessutti IT, et al. Identification and characterization of the GmRD26 soybean promoter in response to abiotic stresses: potential tool for biotechnological application [J]. BMC Biotechnol, 2019, 19(1): 79. |
| [37] | Pimenta MR, Silva PA, Mendes GC, et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence [J]. Plant Cell Physiol, 2016, 57(5): 1098-1114. |
| [38] | Wu YY, Liu XF, Fu BL, et al. Methyl jasmonate enhances ethylene synthesis in kiwifruit by inducing NAC genes that activate ACS1 [J]. J Agric Food Chem, 2020, 68(10): 3267-3276. |
| [39] | 曲硕, 刘芳, 孙浩文, 等. 大豆NAC转录因子生物信息学分析及GmNAC-1克隆和亚细胞定位 [J]. 大豆科学, 2024, 43(5): 523-538. |
| Qu S, Liu F, Sun HW, et al. Bioinformatics analysis of soybean NAC transcription factor and cloning and subcellular localization of GmNAC-1 [J]. Soybean Sci, 2024, 43(5): 523-538. |
| [1] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [2] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [3] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [4] | ZHAO Qiang, CHEN Si-yu, PENG Fang-li, WANG Can, GAO Jie, ZHOU Ling-bo, ZHANG Guo-bing, JIANG Yu-wen, SHAO Ming-bo. Effects of Intercropping and Nitrogen Application on the Diversity and Functions of Soil Bacteria around Sorghum Rhizosphere [J]. Biotechnology Bulletin, 2025, 41(6): 307-316. |
| [5] | TAN Yu-rong, CHEN Dong-liang, YANG Shou-zhen, LAI Zhen-guang, TANG Xiang-min, SUN Zu-dong, ZENG Wei-ying. Functioal Analysis on GmKTI1-like Gene of Soybean Resistance to Bean Pyralid (Lamprosema indicata) [J]. Biotechnology Bulletin, 2025, 41(6): 99-108. |
| [6] | WANG Miao-miao, ZHAO Xiang-long, WANG Zhao-ming, LIU Zhi-peng, YAN Long-feng. Identification of TCP Gene Family in Medicago ruthenica and Their Expression Pattern Analysis under Drought Stress [J]. Biotechnology Bulletin, 2025, 41(6): 179-190. |
| [7] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [8] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [9] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| [10] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [11] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| [12] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [13] | SUN Tian-guo, YI Lan, QIN Xu-yang, QIAO Meng-xue, GU Xin-ying, HAN Yi, SHA Wei, ZHANG Mei-juan, MA Tian-yi. Genome-wide Identification of the DABB Gene Family in Brassica rapa ssp. pekinensis and Expression Analysis under Saline and Alkali Stress [J]. Biotechnology Bulletin, 2025, 41(4): 156-165. |
| [14] | WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.) [J]. Biotechnology Bulletin, 2025, 41(4): 166-175. |
| [15] | HUANG Jin-heng, HUANG Xi, ZHANG Jia-yan, ZHOU Xin-yu, LIAO Pei-ran, YANG Quan. Identification and Expression Analysis of the C3H Gene Family in Grona styracifolia across Different Varieties [J]. Biotechnology Bulletin, 2025, 41(4): 243-255. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||