








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 119-129.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1252
XU Hui-zhen1( ), SHANTWANA Ghimire2, RAJU Kharel3, YUE Yun4, SI Huai-jun1, TANG Xun1(
), SHANTWANA Ghimire2, RAJU Kharel3, YUE Yun4, SI Huai-jun1, TANG Xun1( )
)
Received:2024-12-24
Online:2025-06-26
Published:2025-06-30
Contact:
TANG Xun
E-mail:xuhz@st.gsau.edu.cn;tangxun@gsau.edu.cn
XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1[J]. Biotechnology Bulletin, 2025, 41(6): 119-129.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Purpose |
|---|---|---|
| pCAMBIA1300-StSIZ1-F | ggtacccggggatcctctagaATGGATTTGGTTGCTAGCTGCA | 基因克隆 |
| pCAMBIA1300-StSIZ1-R | gcccttgctcaccatgtcgacCTATTCAGAATCCGAATCAATACTTAGAT | |
| qPCR-StSIZ1-F | TTAGACTTGTGAAGCGGCGT | RT-qPCR |
| qPCR-StSIZ1-R | CCACCAACACAACGACGAAC | |
| Eflα-F | GATGGTCAGACCCGTGAACA | 内参基因 |
| Eflα-R | CCTTGGAGTACTTCGGGGTC |
Table 1 Information about the primers
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Purpose |
|---|---|---|
| pCAMBIA1300-StSIZ1-F | ggtacccggggatcctctagaATGGATTTGGTTGCTAGCTGCA | 基因克隆 |
| pCAMBIA1300-StSIZ1-R | gcccttgctcaccatgtcgacCTATTCAGAATCCGAATCAATACTTAGAT | |
| qPCR-StSIZ1-F | TTAGACTTGTGAAGCGGCGT | RT-qPCR |
| qPCR-StSIZ1-R | CCACCAACACAACGACGAAC | |
| Eflα-F | GATGGTCAGACCCGTGAACA | 内参基因 |
| Eflα-R | CCTTGGAGTACTTCGGGGTC |

Fig. 5 Protein structure analysis of StSIZ1 and StSIZ2 and cloning of StSIZ1 geneA: Conserved structural domains of the StSIZ1 and StSIZ2 protein. B-C: Tertiary structure of StSIZ1 and StSIZ2. D: PCR production of gene StSIZ1
基因ID Gene ID | 基因名 Gene name | 理论分子量 Molecular weight (Da) | 等电点 Point isoelectric | 不稳定指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 亲水度 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| Soltu.DM.11G022540.5 | StSIZ1 | 95 449.87 | 4.96 | 44.28 | 76.34 | -0.453 |
| Soltu.DM.06G005690.1 | StSIZ2 | 93 832.16 | 5.44 | 47.05 | 80.67 | -0.373 |
| Soltu.DM.07G023970.1 | StMMS21 | 28 532.31 | 5.10 | 58.06 | 77.34 | -0.533 |
| Soltu.DM.08G004520.1 | StPIAS1 | 96 099.04 | 6.01 | 48.14 | 73.54 | -0.390 |
| Soltu.DM.06G005850.1 | StPIAS2 | 163 534.42 | 6.74 | 41.34 | 88.45 | -0.296 |
| Soltu.DM.05G011900.1 | StRanBP2a | 30 458.93 | 9.24 | 58.23 | 29.93 | -0.922 |
| Soltu.DM.12G024390.1 | StRanBP2b | 26 349.84 | 8.17 | 52.64 | 58.67 | -0.482 |
| Soltu.DM.09G019740.1 | StRanGAP1a | 58 094.47 | 4.58 | 37.95 | 94.34 | -0.283 |
| Soltu.DM.01G024550.1 | StRanGAP1b | 60 594.10 | 4.64 | 41.86 | 90.05 | -0.386 |
| Soltu.DM.05G007610.1 | StRanGAP1c | 64 532.83 | 8.78 | 30.85 | 99.40 | -0.067 |
Table 2 Physicochemical properties of protein members in the potato SUMO E3 ligase gene family
基因ID Gene ID | 基因名 Gene name | 理论分子量 Molecular weight (Da) | 等电点 Point isoelectric | 不稳定指数 Instability index | 脂肪族氨基酸指数 Aliphatic index | 亲水度 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| Soltu.DM.11G022540.5 | StSIZ1 | 95 449.87 | 4.96 | 44.28 | 76.34 | -0.453 |
| Soltu.DM.06G005690.1 | StSIZ2 | 93 832.16 | 5.44 | 47.05 | 80.67 | -0.373 |
| Soltu.DM.07G023970.1 | StMMS21 | 28 532.31 | 5.10 | 58.06 | 77.34 | -0.533 |
| Soltu.DM.08G004520.1 | StPIAS1 | 96 099.04 | 6.01 | 48.14 | 73.54 | -0.390 |
| Soltu.DM.06G005850.1 | StPIAS2 | 163 534.42 | 6.74 | 41.34 | 88.45 | -0.296 |
| Soltu.DM.05G011900.1 | StRanBP2a | 30 458.93 | 9.24 | 58.23 | 29.93 | -0.922 |
| Soltu.DM.12G024390.1 | StRanBP2b | 26 349.84 | 8.17 | 52.64 | 58.67 | -0.482 |
| Soltu.DM.09G019740.1 | StRanGAP1a | 58 094.47 | 4.58 | 37.95 | 94.34 | -0.283 |
| Soltu.DM.01G024550.1 | StRanGAP1b | 60 594.10 | 4.64 | 41.86 | 90.05 | -0.386 |
| Soltu.DM.05G007610.1 | StRanGAP1c | 64 532.83 | 8.78 | 30.85 | 99.40 | -0.067 |

Fig. 6 Subcellular localization of potato StSIZ1 in the tobacco cellspEGFP: Control (pCAMBIA1300-35S-EGFP vector); pEGFP-StSIZ1: pEGFP-StSIZ1 fusion protein. Scale bar=20 μm
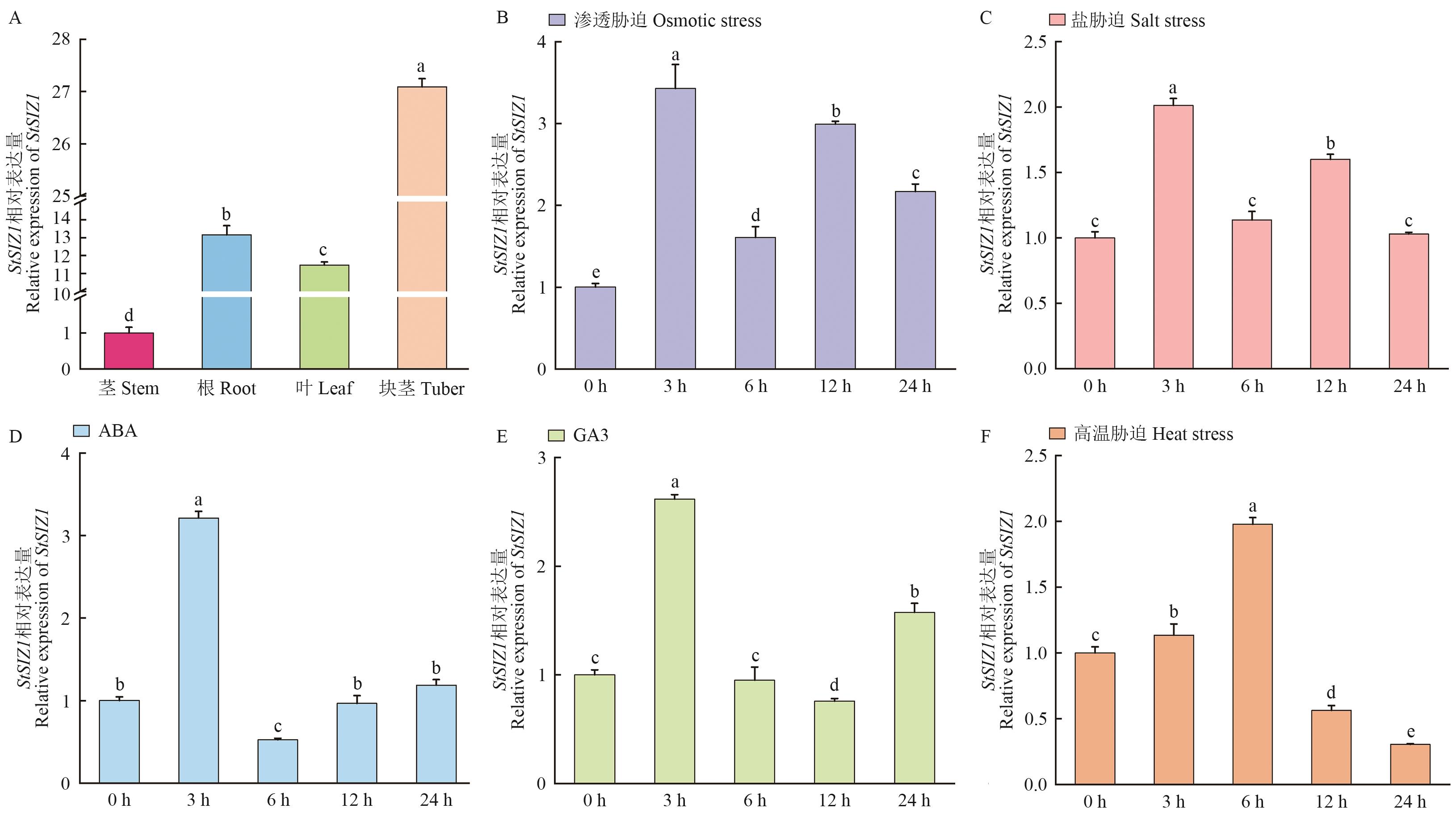
Fig. 7 Expression pattern of StSIZ1 in the potatoA: Expression of StSIZ1 gene in different tissues. Relative expressions of the StSIZ1 under treatment with 20% PEG (B), 150 mmol/L NaCl (C), 50 μmol/LABA (D), 50 μmol/L GA3 (E), and heat stress (35 ℃) (F). Different letters indcate significant differences (P<0.05)
| 1 | Scaramagli S, Biondi S, Leone A, et al. Acclimation to low water potential in potato cell suspension cultures leads to changes in putrescine metabolism [J]. Plant Physiol Biochem, 2000, 38(4): 345-351. |
| 2 | 贾明飞, 樊建英, 封志明, 等. 氮素不同形态对早熟马铃薯产量和氮素积累的影响 [J]. 中国土壤与肥料, 2024, (2): 146-151. |
| Jia MF, Fan JY, Feng ZM, et al. Effects of different forms of nitrogen on yield and nitrogen accumulation in early maturing potatoes [J]. Soil and Fertiliser China, 2024, (2): 146-151. | |
| 3 | 汪昕, 杨德秋, 李洋, 等. 马铃薯机械化收获技术进展与展望 [J]. 农机化研究, 2024, 46(9): 1-7. |
| Wang X, Yang DQ, Li Y, et al. Progress and prospect of potato mechanized harvesting technology [J]. J Agric Mech Res, 2024, 46(9): 1-7. | |
| 4 | Seo J, Lee KJ. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches [J]. J Biochem Mol Biol, 2004, 37(1): 35-44. |
| 5 | Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles [J]. Gene, 2000, 248(1/2): 1-14. |
| 6 | Kurepa J, Walker JM, Smalle J, et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and-2 conjugates is increased by stress [J]. J Biol Chem, 2003, 278(9): 6862-6872. |
| 7 | Elrouby N. Analysis of small ubiquitin-like modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development [J]. Plant Physiol, 2015, 169(2): 1006-1017. |
| 8 | Park HJ, Yun DJ. SUMO proteins grapple with biotic and abiotic stresses in Arabidopsis [J]. J Plant Biol, 2013, 56(2): 77-84. |
| 9 | Novatchkova M, Tomanov K, Hofmann K, et al. Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison [J]. New Phytol, 2012, 195(1): 23-31. |
| 10 | Han YF, Zhao QY, Dang LL, et al. The SUMO E3 ligase-like proteins PIAL1 and PIAL2 interact with MOM1 and form a novel complex required for transcriptional silencing [J]. Plant Cell, 2016, 28(5): 1215-1229. |
| 11 | Ritterhoff T, Das H, Hofhaus G, et al. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes [J]. Nat Commun, 2016, 7: 11482. |
| 12 | Zheng Z, Liu D. SIZ1 regulates phosphate deficiency-induced inhibition of primary root growth of Arabidopsis by modulating Fe accumulation and ROS production in its roots [J]. Plant Signal Behav, 2021, 16(10): 1946921. |
| 13 | Rawat SS, Sandhya S, Laxmi A. Complex genetic interaction between glucose sensor HXK1 and E3 SUMO ligase SIZ1 in regulating plant morphogenesis [J]. Plant Signal Behav, 2024, 19(1): 2341506. |
| 14 | Gao SJ, Zeng XQ, Wang JH, et al. Arabidopsis SUMO E3 ligase SIZ1 interacts with HDA6 and negatively regulates HDA6 function during flowering [J]. Cells, 2021, 10(11): 3001. |
| 15 | Castro PH, Couto D, Santos MÂ, et al. SUMO E3 ligase SIZ1 connects sumoylation and reactive oxygen species homeostasis processes in Arabidopsis [J]. Plant Physiol, 2022, 189(2): 934-954. |
| 16 | Miura K, Nozawa R. Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana [J]. Plant Biotechnol, 2014, 31(2): 167-172. |
| 17 | Kim JY, Song JT, Seo HS. Post-translational modifications of Arabidopsis E3 SUMO ligase AtSIZ1 are controlled by environmental conditions [J]. FEBS Open Bio, 2017, 7(10): 1622-1634. |
| 18 | Yoo CY, Miura K, Jin JB, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid [J]. Plant Physiol, 2006, 142(4): 1548-1558. |
| 19 | Miura K, Sato A, Ohta M, et al. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2 [J]. Planta, 2011, 234(6): 1191-1199. |
| 20 | Jiang H, Zhou LJ, Gao HN, et al. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple [J]. Plant Physiol, 2022, 189(4): 2044-2060. |
| 21 | 张亚丽. 苹果SUMO E3连接酶MdSIZ1蛋白SUMO化MdMYB30调控蜡质形成的机理 [D]. 泰安: 山东农业大学, 2023. |
| Zhang YL. Mechanism of regulating wax formation by sumoylation of apple SUMO E3 ligase MdSIZ1 protein MdMYB30 [D]. Tai'an: Shandong Agricultural University, 2023. | |
| 22 | Zhang S, Wang SJ, Lv JL, et al. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato [J]. Plant Cell Physiol, 2018, 59(1): 58-71. |
| 23 | Zhang S, Zhuang KY, Wang SJ, et al. A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco [J]. J Integr Plant Biol, 2017, 59(2): 102-117. |
| 24 | Cai B, Kong XX, Zhong C, et al. SUMO E3 Ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean [J]. J Integr Plant Biol, 2017, 59(1): 2-14. |
| 25 | Li YJ, Wang GX, Xu ZQ, et al. Organization and regulation of soybean SUMOylation system under abiotic stress conditions [J]. Front Plant Sci, 2017, 8: 1458. |
| 26 | Lei SK, Wang QZ, Chen Y, et al. Capsicum SIZ1 contributes to ABA-induced SUMOylation in pepper [J]. Plant Sci, 2022, 314: 111099. |
| 27 | Lai RQ, Jiang JM, Wang J, et al. Functional characterization of three maize SIZ/PIAS-type SUMO E3 ligases [J]. J Plant Physiol, 2022, 268: 153588. |
| 28 | 汪永平. 梭梭SUMO E3连接酶基因HaSIZ1的克隆和功能分析 [D]. 兰州: 兰州大学, 2020. |
| Wang YP. Cloning and functional analysis of HaSIZ1 ligase gene of Haloxylon ammodendron SUMO E3 [D]. Lanzhou: Lanzhou University, 2020. | |
| 29 | Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets [J]. Mol Biol Evol, 2016, 33(7): 1870-1874. |
| 30 | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| 31 | Srivastava M, Verma V, Srivastava AK. The converging path of protein SUMOylation in phytohormone signalling: highlights and new frontiers [J]. Plant Cell Rep, 2021, 40(11): 2047-2061. |
| 32 | Crozet P, Margalha L, Butowt R, et al. SUMOylation represses SnRK1 signaling in Arabidopsis [J]. Plant J, 2016, 85(1): 120-133. |
| 33 | Xiong JW, Yang FB, Wei F, et al. Inhibition of SIZ1-mediated SUMOylation of HOOKLESS1 promotes light-induced apical hook opening in Arabidopsis [J]. Plant Cell, 2023, 35(6): 2027-2043. |
| 34 | Cohen-Peer R, Schuster S, Meiri D, et al. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance [J]. Plant Mol Biol, 2010, 74(1/2): 33-45. |
| 35 | Garcia-Dominguez M, March-Diaz R, Reyes JC. The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins [J]. J Biol Chem, 2008, 283(31): 21469-21477. |
| 36 | Suzuki R, Shindo H, Tase A, et al. Solution structures and DNA binding properties of the N-terminal SAP domains of SUMO E3 ligases from Saccharomyces cerevisiae and Oryza sativa [J]. Proteins, 2009, 75(2): 336-347. |
| 37 | Catala R, Ouyang J, Abreu IA, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses [J]. Plant Cell, 2007, 19(9): 2952-2966. |
| 38 | 赵媛媛, 刘萍涛, 康建宏. 高温胁迫对马铃薯叶片生理特性及产量影响研究 [J]. 中国农学通报, 2024, 40(21): 35-44. |
| Zhao YY, Liu PT, Kang JH. Effects of high temperature stress on physiological characteristics and yield of potato leaves [J]. Chin Agric Sci Bull, 2024, 40(21): 35-44. | |
| 39 | Wyrzykowska A, Bielewicz D, Plewka P, et al. The MYB33, MYB65, and MYB101 transcription factors affect Arabidopsis and potato responses to drought by regulating the ABA signaling pathway [J]. Physiol Plant, 2022, 174(5): e13775. |
| [1] | LUO Ji-lin, LI Jin-ye, JIA Yu-xin. Identification and Functional Analysis of Gravity Response Regulatory Genes in Potato [J]. Biotechnology Bulletin, 2025, 41(6): 109-118. |
| [2] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [3] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [4] | LIU Xin, WANG Jia-wen, LI Jin-wei, MOU Ce, YANG Pan-pan, MING Jun, XU Lei-feng. Cloning and Expression Analysis of Three LdBBXs in Lilium davidii var. willmottiae [J]. Biotechnology Bulletin, 2025, 41(5): 186-196. |
| [5] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [6] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [7] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| [8] | WEN Bo-lin, WAN Min, HU Jian-jun, WANG Ke-xiu, JING Sheng-lin, WANG Xin-yue, ZHU Bo, TANG Ming-xia, LI Bing, HE Wei, ZENG Zi-xian. Establishment of Genetic Transformation and Gene Editing System for a Potato Cultivar Chuanyu 50 [J]. Biotechnology Bulletin, 2025, 41(4): 88-97. |
| [9] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [10] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [11] | YU Ting, HUANG Dan-dan, ZHU Yan-hui, YANG Mei-hong, AI Ju, GAO Dong-li. Screening and Interaction Verification of Transcription Factors Stpatatin 05 Gene in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 137-145. |
| [12] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [13] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [14] | PENG Ting, LIN Ying, TAN Yuan-yuan, RAO Ying, HUANG Qin, ZHANG Wen-e, WANG Bo, TIAN Rui-feng, LIU Guo-feng. Cloning and Expression Analysis of AwANS Genes in Allium wallichii [J]. Biotechnology Bulletin, 2025, 41(3): 230-239. |
| [15] | XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3 [J]. Biotechnology Bulletin, 2025, 41(2): 150-162. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||