








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 289-301.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0215
LI Ya-tao1( ), ZHANG Zhi-peng2,3, ZHAO Meng-yao1, LYU Zhen1, GAN Tian1, WEI Hao3, WU Shu-feng3, MA Yu-chao1(
), ZHANG Zhi-peng2,3, ZHAO Meng-yao1, LYU Zhen1, GAN Tian1, WEI Hao3, WU Shu-feng3, MA Yu-chao1( )
)
Received:2025-03-02
Online:2025-09-26
Published:2025-09-24
Contact:
MA Yu-chao
E-mail:lyt888888@bjfu.edu.cn;mayuchao@bifu.edu.cn
LI Ya-tao, ZHANG Zhi-peng, ZHAO Meng-yao, LYU Zhen, GAN Tian, WEI Hao, WU Shu-feng, MA Yu-chao. Whole Genome Analysis of Bradyrhizobium sp. Bd1 and the Negative Regulating Function of TetR3 during Cell Growth and Nodulation[J]. Biotechnology Bulletin, 2025, 41(9): 289-301.

Fig. 1 Construction of TetR3-inactive mutantA: Construction of recombinant plasmid and conjugation donor. B: TetR3-inactive result. C: Sequencing of the mutant. D: Colony PCR verification (M: DNA marker 2000; lane 1: Bd1; lane 2: Bd1ΔTetR3)
| Family | Gene ID | Name | KEGG gene ID | KO description | Location |
|---|---|---|---|---|---|
| fix | gene5832 | fixK | bja:bll3466 | CRP/FNR family transcriptional regulator | 6070103-6070786 |
| gene6542 | fixK | bja:bll2757 | CRP/FNR family transcriptional regulator | 6868843-6869541 | |
| gene6539 | fixL | bja:bll2760 | LuxR family, sensor kinase FixL | 6866141-6867658 | |
| gene6540 | fixJ | bja:bll2759 | LuxR family, response regulator FixJ | 6867662-6868279 | |
| gene7359 | fixA | bja:blr2038 | Flavoprotein beta subunit | 7803669-7804535 | |
| gene7895 | fixA | bja:blr1377 | Flavoprotein beta subunit | 8425108-8425857C | |
| gene7566 | fixX | bja:bsr1775 | Ferredoxin like protein | 8067228-8067524C | |
| gene7567 | fixC | bja:blr1774 | Flavoprotein-quinone oxidoreductase | 8067563-8068870C | |
| gene7568 | fixB | bja:blr1773 | Flavoprotein alpha subunit | 8068882-8069991C | |
| gene7894 | fixB | bja:blr1378 | Flavoprotein alpha subunit | 8424164-8425108C | |
| nod | gene5597 | nodT | bsym:CIT39_22430 | Multidrug efflux system | 5805752-5807308 |
| gene7363 | nodU | bja:blr2034 | Carbamoyltransferase | 7809897-7811504C | |
| gene7367 | nodU | bjp:RN69_37980 | Carbamoyltransferase | 7813893-7815602C | |
| gene7365 | nodJ | bja:blr2031 | Lipooligosaccharide transport system permease | 7812179-7812967C | |
| gene7366 | nodI | bja:blr2030 | Lipooligosaccharide transport system ATP-binding protein | 7812971-7813891C | |
| gene7368 | nodC | bja:blr2027 | N-acetylglucosaminyltransferase | 7816172-7817629C | |
| gene7369 | nodB | bja:blr2026 | Chitooligosaccharide deacetylase | 7817644-7818303C | |
| gene7370 | nodA | bja:blr2025 | Nodulation protein A | 7818300-7818932C | |
| gene7372 | nodD | bja:bll2023 | LysR family transcriptional regulator | 7819726-7820670 | |
| nif | gene7360 | nifA | bja:blr2037 | Nif-specific regulatory protein | 7805053-7806801C |
| gene7569 | nifW | bja:blr1771 | Nitrogenase-stabilizing/protective protein | 8070942-8071283C | |
| gene7571 | nifQ | bju:BJ6T_80490 | Nitrogen fixation protein NifQ | 8071938-8072714C | |
| gene7572 | nifH | bja:blr1769 | Nitrogenase iron protein NifH | 8072861-8073745 | |
| gene7580 | nifZ | bjp:RN69_39140 | Nitrogen fixation protein NifZ | 8078154-8078468C | |
| gene7582 | nifB | bju:BJ6T_80610 | Nitrogen fixation protein NifB | 8078954-8080537C | |
| gene7583 | nifT | bja:bsr1757 | Nitrogen fixation protein NifT | 8081201-8081425C | |
| gene7594 | nifX | bja:blr1747 | Nitrogen fixation protein NifX | 8090471-8090866C | |
| gene7595 | nifN | bja:blr1746 | Nitrogenase molybdenum-iron protein NifN | 8090863-8092272C | |
| gene7596 | nifE | bja:blr1745 | Nitrogenase molybdenum-cofactor synthesis protein NifE | 8092282-8093925C | |
| gene7597 | nifK | bja:blr1744 | Molybdenum-iron protein beta chain | 8094018-8095574C | |
| gene7598 | nifD | bja:blr1743 | Molybdenum-iron protein alpha chain | 8095640-8097142C |
Table 1 Nitrogen fixation-related genes in Bd1 genome
| Family | Gene ID | Name | KEGG gene ID | KO description | Location |
|---|---|---|---|---|---|
| fix | gene5832 | fixK | bja:bll3466 | CRP/FNR family transcriptional regulator | 6070103-6070786 |
| gene6542 | fixK | bja:bll2757 | CRP/FNR family transcriptional regulator | 6868843-6869541 | |
| gene6539 | fixL | bja:bll2760 | LuxR family, sensor kinase FixL | 6866141-6867658 | |
| gene6540 | fixJ | bja:bll2759 | LuxR family, response regulator FixJ | 6867662-6868279 | |
| gene7359 | fixA | bja:blr2038 | Flavoprotein beta subunit | 7803669-7804535 | |
| gene7895 | fixA | bja:blr1377 | Flavoprotein beta subunit | 8425108-8425857C | |
| gene7566 | fixX | bja:bsr1775 | Ferredoxin like protein | 8067228-8067524C | |
| gene7567 | fixC | bja:blr1774 | Flavoprotein-quinone oxidoreductase | 8067563-8068870C | |
| gene7568 | fixB | bja:blr1773 | Flavoprotein alpha subunit | 8068882-8069991C | |
| gene7894 | fixB | bja:blr1378 | Flavoprotein alpha subunit | 8424164-8425108C | |
| nod | gene5597 | nodT | bsym:CIT39_22430 | Multidrug efflux system | 5805752-5807308 |
| gene7363 | nodU | bja:blr2034 | Carbamoyltransferase | 7809897-7811504C | |
| gene7367 | nodU | bjp:RN69_37980 | Carbamoyltransferase | 7813893-7815602C | |
| gene7365 | nodJ | bja:blr2031 | Lipooligosaccharide transport system permease | 7812179-7812967C | |
| gene7366 | nodI | bja:blr2030 | Lipooligosaccharide transport system ATP-binding protein | 7812971-7813891C | |
| gene7368 | nodC | bja:blr2027 | N-acetylglucosaminyltransferase | 7816172-7817629C | |
| gene7369 | nodB | bja:blr2026 | Chitooligosaccharide deacetylase | 7817644-7818303C | |
| gene7370 | nodA | bja:blr2025 | Nodulation protein A | 7818300-7818932C | |
| gene7372 | nodD | bja:bll2023 | LysR family transcriptional regulator | 7819726-7820670 | |
| nif | gene7360 | nifA | bja:blr2037 | Nif-specific regulatory protein | 7805053-7806801C |
| gene7569 | nifW | bja:blr1771 | Nitrogenase-stabilizing/protective protein | 8070942-8071283C | |
| gene7571 | nifQ | bju:BJ6T_80490 | Nitrogen fixation protein NifQ | 8071938-8072714C | |
| gene7572 | nifH | bja:blr1769 | Nitrogenase iron protein NifH | 8072861-8073745 | |
| gene7580 | nifZ | bjp:RN69_39140 | Nitrogen fixation protein NifZ | 8078154-8078468C | |
| gene7582 | nifB | bju:BJ6T_80610 | Nitrogen fixation protein NifB | 8078954-8080537C | |
| gene7583 | nifT | bja:bsr1757 | Nitrogen fixation protein NifT | 8081201-8081425C | |
| gene7594 | nifX | bja:blr1747 | Nitrogen fixation protein NifX | 8090471-8090866C | |
| gene7595 | nifN | bja:blr1746 | Nitrogenase molybdenum-iron protein NifN | 8090863-8092272C | |
| gene7596 | nifE | bja:blr1745 | Nitrogenase molybdenum-cofactor synthesis protein NifE | 8092282-8093925C | |
| gene7597 | nifK | bja:blr1744 | Molybdenum-iron protein beta chain | 8094018-8095574C | |
| gene7598 | nifD | bja:blr1743 | Molybdenum-iron protein alpha chain | 8095640-8097142C |
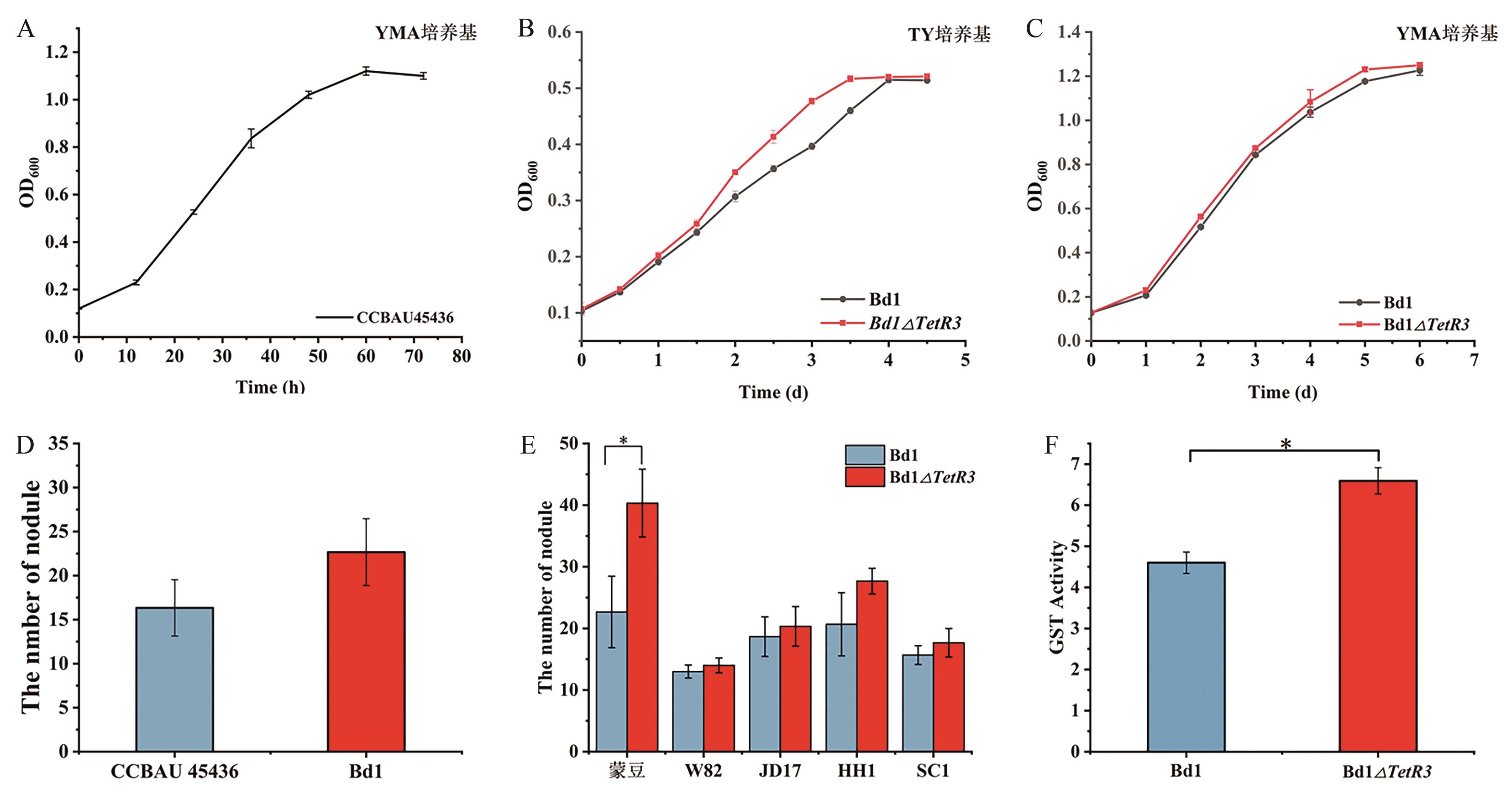
Fig. 7 Functional characteristics of Bd1 and the mutant Bd1ΔTetR3A: Growth curve of S. fredii CCBAU45436 in YMA medium. B and C: Growth of the mutant Bd1ΔTetR3 in YMA and TY, respectively. D: Number of symbiotic nodules of Bd1 and S. fredii CCBAU 45436 with Glycine max Mengdou 1137; E: Number of symbiotic nodules between mutant and different soybean varieties. F: Glutathione S-transferase activity of the mutant Bd1ΔTetR3. *P<0.05
| [1] | 向星宇, 吴雪许, 许建双, 等. 中国大豆进口贸易现状及对策研究 [J]. 黑龙江粮食, 2024, (11): 26-28. |
| Xiang XY, Wu XX, Xu JS, et al. China's soybean import trade: current status and policy recommendations [J]. Heilongjiang Grain, 2024, (11): 26-28. | |
| [2] | 王善明. 华癸中慢生根瘤菌7653R全基因组测序及比较基因组学研究 [D]. 武汉: 华中农业大学, 2014. |
| Wang SM. Whole-genome sequencing and comparative genomic analysis of Mesorhizobium huakuii strain7653R [D]. Wuhan: Huazhong Agricultural University, 2014. | |
| [3] | Margaret I, Becker A, Blom J, et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean [J]. J Biotechnol, 2011, 155(1): 11-19. |
| [4] | Yang SH, Chen WH, Wang ET, et al. Rhizobial biogeography and inoculation application to soybean in four regions across China [J]. J Appl Microbiol, 2018, 125(3): 853-866. |
| [5] | Tian CF, Zhou YJ, Zhang YM, et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations [J]. Proc Natl Acad Sci USA, 2012, 109(22): 8629-8634. |
| [6] | Balhana RJC, Singla A, Sikder MH, et al. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions [J]. BMC Genomics, 2015, 16(1): 479. |
| [7] | Ramos JL, Martínez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors [J]. Microbiol Mol Biol Rev, 2005, 69(2): 326-356. |
| [8] | 吴攀攀, 李博文, 陈克涛, 等. TetR家族转录调控因子配体的研究进展 [J]. 生物工程学报, 2021, 37(7): 2379-2392. |
| Wu PP, Li BW, Chen KT, et al. Ligands of TetR family transcriptional regulators: a review [J]. Chin J Biotechnol, 2021, 37(7): 2379-2392. | |
| [9] | Patil RS, Sharma S, Bhaskarwar AV, et al. TetR and OmpR family regulators in natural product biosynthesis and resistance [J]. Proteins, 2025, 93(1): 38-71. |
| [10] | Ohkama-Ohtsu N, Honma H, Nakagome M, et al. Growth Rate of and Gene Expression in Bradyrhizobium diazoefficiens USDA110 due to a Mutation in blr7984, a TetR Family Transcriptional Regulator Gene [J]. Microbes Environ, 2016, 31(3): 249-259. |
| [11] | Han F, He XQ, Chen WW, et al. Involvement of a novel TetR-like regulator (BdtR) of Bradyrhizobium diazoefficiens in the efflux of isoflavonoid genistein [J]. Mol Plant Microbe Interact, 2020, 33(12): 1411-1423. |
| [12] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler [J]. Gigascience, 2012, 1(1): 18. |
| [13] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics [J]. Genome Res, 2009, 19(9): 1639-1645. |
| [14] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013 [J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [15] | Zhang XX, Guo HJ, Jiao J, et al. Pyrosequencing of rpoB uncovers a significant biogeographical pattern of rhizobial species in soybean rhizosphere [J]. J Biogeogr, 2017, 44(7): 1491-1499. |
| [16] | 张星星. 大豆慢生根瘤菌分子进化学分析及土著大豆根瘤菌生物地理分布的高通量研究 [D]. 北京: 中国农业大学, 2014. |
| Zhang XX. Molecular evolution of soybean nodulating Bradyrhizobium and biogeography of indigenous soybean rhizobia revealed by amplicon pyrosequencing [D]. Beijing: China Agricultural University, 2014. | |
| [17] | Wang CH, Li M, Zhao Y, et al. SHORT-ROOT paralogs mediate feedforward regulation of D-type cyclin to promote nodule formation in soybean [J]. Proc Natl Acad Sci USA, 2022, 119(3): e2108641119. |
| [18] | Bender FR, Nagamatsu ST, Delamuta JRM, et al. Genetic variation in symbiotic islands of natural variant strains of soybean Bradyrhizobium japonicum and Bradyrhizobium diazoefficiens differing in competitiveness and in the efficiency of nitrogen fixation [J]. Microb Genom, 2022, 8(4): 000795. |
| [19] | Colclough AL, Scadden J, Blair JA. TetR-family transcription factors in Gram-negative bacteria: conservation, variation and implications for efflux-mediated antimicrobial resistance [J]. BMC Genomics, 2019, 20(1): 731. |
| [20] | Singh P, Jain A, Chhabra R, et al. TetR family transcriptional regulators: Lipid metabolism and drug resistance in mycobacteria [J]. Gene Rep, 2024, 36: 101938. |
| [21] | Henríquez T, Stein NV, Jung H. Resistance to bipyridyls mediated by the TtgABC efflux system in Pseudomonas putida KT2440 [J]. Front Microbiol, 2020, 11: 1974. |
| [22] | Watanabe S, Shimada N, Tajima K, et al. Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism. Novel evolutionary insight into sugar metabolism [J]. J Biol Chem, 2006, 281(44): 33521-33536. |
| [23] | Nguyen Le Minh P, de Cima S, Bervoets I, et al. Ligand binding specificity of RutR, a member of the TetR family of transcription regulators in Escherichia coli [J]. FEBS Open Bio, 2015, 5: 76-84. |
| [24] | Kendall SL, Burgess P, Balhana R, et al. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2 [J]. Microbiology, 2010, 156(Pt 5): 1362-1371. |
| [25] | Erni B, Siebold C, Christen S, et al. Small substrate, big surprise: fold, function and phylogeny of dihydroxyacetone kinases [J]. Cell Mol Life Sci, 2006, 63(7/8): 890-900. |
| [26] | Li YJ, Wang CH, Zheng L, et al. Natural variation of GmRj2/Rfg1 determines symbiont differentiation in soybean [J]. Curr Biol, 2023, 33(12): 2478-2490.e5. |
| [27] | Wani SR, Dubey AA, Jain V. Ms6244 is a novel Mycobacterium smegmatis TetR family transcriptional repressor that regulates cell growth and morphophysiology [J]. FEBS Lett, 2023, 597(10): 1428-1440. |
| [28] | Lei YK, Asamizu S, Ishizuka T, et al. Regulation of multidrug efflux pumps by TetR family transcriptional repressor negatively affects secondary metabolism in Streptomyces coelicolor A3(2) [J]. Appl Environ Microbiol, 2023, 89(3): e0182222. |
| [29] | Schmacht M, Lorenz E, Senz M. Microbial production of glutathione [J]. World J Microbiol Biotechnol, 2017, 33(6): 106. |
| [30] | Luo S, Yin J, Peng Y, et al. Glutathione is involved in detoxification of peroxide and root nodule symbiosis of Mesorhizobium huakuii [J]. Curr Microbiol, 2020, 77(1): 1-10. |
| [31] | Van Laar TA, Esani S, Birges TJ, et al. Pseudomonas aeruginosa gshA mutant is defective in biofilm formation, swarming, and pyocyanin production [J]. mSphere, 2018, 3(2): e00155-18. |
| [32] | Ku JWK, Gan YH. New roles for glutathione: Modulators of bacterial virulence and pathogenesis [J]. Redox Biol, 2021, 44: 102012. |
| [1] | ZHANG Yong-yan, GUO Si-jian, LI Jing, HAO Si-yi, LI Rui-de, LIU Jia-peng, CHENG Chun-zhen. Gene Cloning and Functional Analysis of the Anthocyanin-related VcGSTF19 Gene in Blueberry (Vaccinium corymbosum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 139-146. |
| [2] | WU Ze-yin, HUANG Chen-yang, ZHAO Meng-ran, ZHANG Li-jiao, YAO Fang-jie. Genome-specific Analysis of Pleurotus cornucopiae CCMSSC 04611 with Short Stipe [J]. Biotechnology Bulletin, 2025, 41(5): 320-332. |
| [3] | ZHANG Hui, LU Wen-cai, WANG Dong, LIU Qian, MA Lian-jie. Identification of Bacillus cereus YT2-1C with High Indoleacetic Acid Yield and Its Growth-promoting Effect [J]. Biotechnology Bulletin, 2025, 41(5): 300-309. |
| [4] | SONG Ying-pei, WANG Can, ZHOU Hui-wen, KONG Ke-ke, XU Meng-ge, WANG Rui-kai. Analysis of Soybean Pod Dehiscence Habit Based on Whole Genome Association Analysis and Genetic Diversity [J]. Biotechnology Bulletin, 2025, 41(2): 97-106. |
| [5] | ZHOU Jiang-hong, XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen. Whole Genome Sequencing and Comparative Genomic Analysis of Antagonistic Bacterium CCBC3-3-1 against Verticillium dahlia [J]. Biotechnology Bulletin, 2024, 40(7): 235-246. |
| [6] | TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain [J]. Biotechnology Bulletin, 2024, 40(7): 323-334. |
| [7] | WANG Zi, SHI Jin-chuan, WANG Yong-qiang, SUN Miao, MENG Ling-hao, GENG Chao, LIU Kai. Whole Genome Sequencing and Genome Evolution Analysis of Capsular Serotype A and D Pasteurella multocida of Bovine [J]. Biotechnology Bulletin, 2024, 40(12): 282-290. |
| [8] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [9] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [10] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [11] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [12] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [13] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [14] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [15] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||