








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 248-256.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1348
收稿日期:2020-11-03
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:刘纹甫,男,硕士研究生,研究方向:分子生物学;E-mail: 基金资助:
LIU Wen-fu( ), HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang(
), HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang( )
)
Received:2020-11-03
Published:2021-11-26
Online:2021-12-03
摘要:
Whirly转录因子广泛存在于植物中,参与植物生长发育进程和逆境胁迫耐受,对其功能的研究有助于我们理解雨生红球藻响应胁迫的机制。因此,先根据雨生红球藻192.80转录组测序数据,分离克隆到一个Whirly转录因子序列,并对其结构及表达进行分析。该转录因子的cDNA序列为924 bp,基因组序列为2 556 bp,由有5个外显子和4个内含子组成,命名为HpWhirly1。HpWhirly1预测由244个氨基酸组成,拥有Whirly转录因子的保守结构域。HpWhirly1在醋酸钠和水杨酸胁迫早期表达上调,在强光-醋酸钠联合胁迫和缺氮胁迫条件下表达下调,表明其在雨生红球藻受到胁迫时发挥一定的作用。以上研究结果将有助于进一步研究Whirly转录因子的功能及其在雨生红球藻响应胁迫时的作用。
刘纹甫, 黄丹琼, 胡群菊, 王潮岗. 雨生红球藻转录因子Whirly1的获得及其表达分析[J]. 生物技术通报, 2021, 37(11): 248-256.
LIU Wen-fu, HUANG Dan-qiong, HU Qun-ju, WANG Chao-gang. Acquisition and Expression Analysis of the Transcription Factor Whirly1 from Haematococcus pluvialis[J]. Biotechnology Bulletin, 2021, 37(11): 248-256.

图3 HpWhirly1基因结构分析 橙色部分为外显子,蓝色部分为UTR区域,黑线为内含子
Fig.3 Gene structure analysis of HpWhirly1 The orange parts are exons,the blue parts are UTR regions,and the black lines are introns

图6 HpWhirly1与其他物种的Whirly同源性分析 Haematococcus lacustris(GFH09517.1),Chlamydomonas eustigma(GAX74163.1),Dunaliella salina(KAF5835724.1),Micractinium conductrix(PSC72124.1);以及模式生物拟南芥中3个Whirly转录因子序列:AtWHY1(AT1G14410),AtWHY2(AT1G71260),AtWHY3(AT2G02740)
Fig.6 Homology analysis of HpWhirly1 and other WHYs from different species Haematococcus lacustris(GFH09517.1),Chlamydomonas eustigma(GAX74163.1),Dunaliella salina(KAF5835724.1),and Micractinium conductrix(PSC72124.1). Three Whirly transcription factor sequences in the model organism Arabidopsis:AtWHY1(AT1G14410),AtWHY2(AT1G71260),and AtWHY3(AT2G02740)
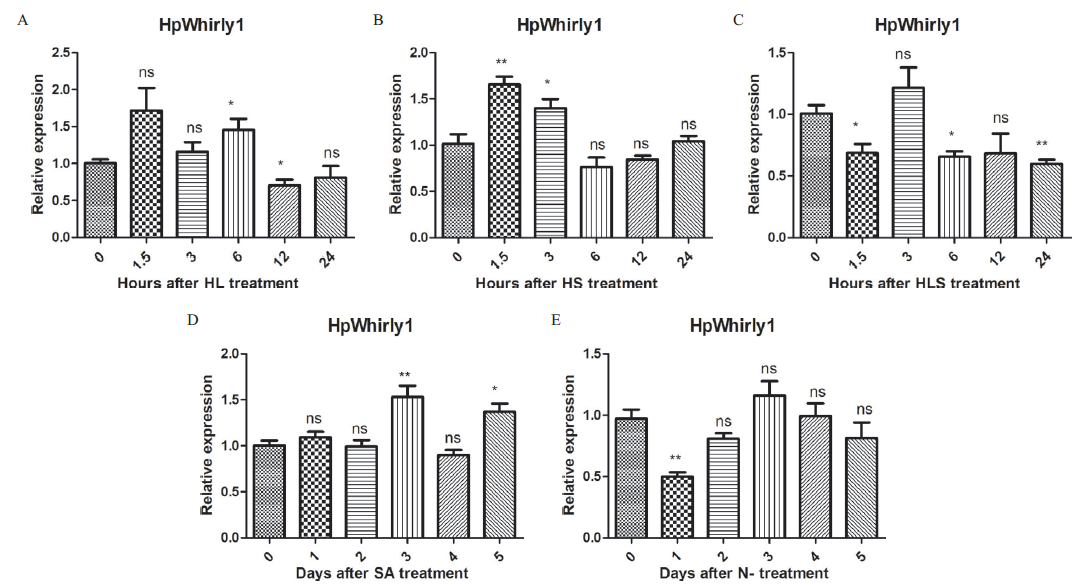
图8 不同胁迫条件下HpWhirly1的表达分析 A:强光处理;B:醋酸处理;C:强光醋酸钠联合处理;D:水杨酸处理;E:缺氮处理。“*”表示处理组和对照组的表达水平存在显著性差异(P<0.05);“**”表示存在极显著性差异(P<0.01)
Fig.8 Expression analysis of HpWhirly1 under different stress conditions A: Strong light treatment. B: Acetic acid treatment. C: Strong light sodium acetate combined treatment. D: Salicylic acid treatment. E: Nitrogen deficiency treatment. * indicates significant difference between treatment group and control group(P<0.05). ** indicates extremely significant difference between treatment group and control group(P<0.01)
| [1] |
Lee YK, Ding SY. Cell cycle and accumulation of astaxanthin in Haematococcus lacustris(chlorophyta)1[J]. J Phycol, 1994, 30(3): 445-449.
doi: 10.1111/j.0022-3646.1994.00445.x URL |
| [2] |
Chamovitz D, Sandmann G, Hirschberg J. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynjournal[J]. J Biol Chem, 1993, 268(23): 17348-17353.
pmid: 8349618 |
| [3] |
Steinbrenner J, Linden H. Regulation of two carotenoid biosynjournal genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis[J]. Plant Physiol, 2001, 125(2): 810-817.
pmid: 11161038 |
| [4] | 王坤鹏. 雨生红球藻的转录组测序及乙酰转移酶基因的克隆和分析[D]. 深圳:深圳大学, 2017. |
| Wang KP. Transcriptome sequencing of haematoccus Pluvialis and cloning and analysis of the acetyltransferase gene[D]. Shenzhen:Shenzhen University, 2017. | |
| [5] | Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses[J]. Biochim Biophys Acta, 2012, 1819(2): 86-96. |
| [6] |
Dickey TH, Altschuler SE, Wuttke DS. Single-stranded DNA-binding proteins:multiple domains for multiple functions[J]. Structure, 2013, 21(7): 1074-1084.
doi: 10.1016/j.str.2013.05.013 URL |
| [7] | 姚沁涛, 张文蔚, 刘莉, 等. Whirly转录因子对非寄主菌诱导水稻HR反应的负调控作用[J]. 中国农业科技导报, 2008, 10(5): 53-58. |
| Yao QT, Zhang WW, Liu L, et al. Negative regulation of rice whirly transcription factor for the hypersensitive response induced by non-host pathogen bacterium[J]. J Agric Sci Technol, 2008, 10(5): 53-58. | |
| [8] | 姚侠妹. 侧柏古树端粒相关基因克隆及其抗逆功能研究[D]. 北京:中国林业科学研究院, 2017. |
| Yao XM. Identification of telomere-related genes from old Platycladus orientalis and their involvement in abiotic stress tolerance[D]. Beijing:Chinese Academy of Forestry, 2017. | |
| [9] |
Zhang YF, Hou MM, Tan BC. The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize[J]. PLoS One, 2013, 8(6): e67369.
doi: 10.1371/journal.pone.0067369 URL |
| [10] |
Isemer R, Krause K, Grabe N, et al. Plastid located WHIRLY1 enhances the responsiveness of Arabidopsis seedlings toward abscisic acid[J]. Front Plant Sci, 2012, 3: 283.
doi: 10.3389/fpls.2012.00283 pmid: 23269926 |
| [11] | 蔡倩, 任育军, 黄晨星, 等. 植物单链DNA结合蛋白WHIRLY2研究进展[J]. 福建农林大学学报:自然科学版, 2018, 47(3): 267-273. |
| Cai Q, Ren YJ, Huang CX, et al. Research progress of single-stranded DNA binding protein WHIRLY2 in plants[J]. J Fujian Agric For Univ:Nat Sci Ed, 2018, 47(3): 267-273. | |
| [12] |
Yoo HH, Kwon C, Lee MM, et al. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis[J]. Plant J, 2007, 49(3): 442-451.
doi: 10.1111/tpj.2007.49.issue-3 URL |
| [13] |
Xiong JY, Lai CX, Qu Z, et al. Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression[J]. Plant Mol Biol, 2009, 71(4/5): 437-449.
doi: 10.1007/s11103-009-9533-7 URL |
| [14] |
Miao Y, Jiang J, Ren Y, et al. The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis[J]. Plant Physiol, 2013, 163(2): 746-756.
doi: 10.1104/pp.113.223412 URL |
| [15] |
Krause K, Kilbienski I, Mulisch M, et al. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles[J]. FEBS Lett, 2005, 579(17): 3707-3712.
doi: 10.1016/j.febslet.2005.05.059 URL |
| [16] |
Desveaux D, Allard J, Brisson N, et al. A new family of plant transcription factors displays a novel ssDNA-binding surface[J]. Nat Struct Biol, 2002, 9(7): 512-517.
pmid: 12080340 |
| [17] |
Desveaux D, Subramaniam R, Després C, et al. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis[J]. Dev Cell, 2004, 6(2): 229-240.
pmid: 14960277 |
| [18] |
Desveaux D, Maréchal A, Brisson N. Whirly transcription factors:defense gene regulation and beyond[J]. Trends Plant Sci, 2005, 10(2): 95-102.
pmid: 15708347 |
| [19] |
Desveaux D, Després C, Joyeux A, et al. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato[J]. Plant Cell, 2000, 12(8): 1477-1489.
pmid: 10948264 |
| [20] | 王洁如. 番茄LeWhy1低温胁迫下的功能分析及转录组分析[D]. 泰安:山东农业大学, 2015. |
| Wang JR. Functional analysis and RNA-sequencing of LeWhy1 in tomato under chilling stress[D]. Tai’an:Shandong Agricultural University, 2015. | |
| [21] | 林文芳, 任育军, 缪颖. 植物Whirly蛋白调控叶片衰老的研究进展[J]. 植物生理学报, 2014, 50(9): 1274-1284. |
| Lin WF, Ren YJ, Miao Y. Research progress of whirly proteins in regulation of leaf senescence[J]. Plant Physiol J, 2014, 50(9): 1274-1284. | |
| [22] |
Cappadocia L, Parent JS, Zampini E, et al. A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage[J]. Nucleic Acids Res, 2012, 40(1): 258-269.
doi: 10.1093/nar/gkr740 pmid: 21911368 |
| [23] |
Grabowski E, Miao Y, Mulisch M, et al. Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell[J]. Plant Physiol, 2008, 147(4): 1800-1804.
doi: 10.1104/pp.108.122796 pmid: 18678751 |
| [24] |
Isemer R, Mulisch M, Schäfer A, et al. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus[J]. FEBS Lett, 2012, 586(1): 85-88.
doi: 10.1016/j.febslet.2011.11.029 pmid: 22154598 |
| [25] |
Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology[J]. Nucleic Acids Res, 2002, 30(6): 1292-1305.
pmid: 11884626 |
| [26] |
Cappadocia L, Sygusch J, Brisson N. Purification, crystallization and preliminary X-ray diffraction analysis of the Whirly domain of StWhy2 in complex with single-stranded DNA[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2008, 64(pt 11): 1056-1059.
doi: 10.1107/S1744309108032399 pmid: 18997341 |
| [27] |
Cappadocia L, Parent JS, Sygusch J, et al. A family portrait:structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2013, 69(pt 11): 1207-1211.
doi: 10.1107/S1744309113028698 pmid: 24192350 |
| [28] |
Cappadocia L, Maréchal A, Parent JS, et al. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair[J]. Plant Cell, 2010, 22(6): 1849-1867.
doi: 10.1105/tpc.109.071399 URL |
| [29] |
Maréchal A, Parent JS, Sabar M, et al. Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function[J]. BMC Plant Biol, 2008, 8: 42.
doi: 10.1186/1471-2229-8-42 URL |
| [30] | 董庆霖, 赵学明, 邢向英. 乙酸钠诱导雨生红球藻合成虾青素的机理[J]. 微生物学通报, 2007, 34(2): 256-260. |
| Dong QL, Zhao XM, Xing XY. Inducing mechanism of acetate on astaxanthin synjournal in Haematococcus pluvialis[J]. Microbiology, 2007, 34(2): 256-260. | |
| [31] |
Gao ZQ, Miao XX, Zhang XW, et al. Comparative fatty acid transcriptomic test and iTRAQ-based proteomic analysis in Haematococcus pluvialis upon salicylic acid(SA)and jasmonic acid(JA)inductions[J]. Algal Res, 2016, 17: 277-284.
doi: 10.1016/j.algal.2016.05.012 URL |
| [32] |
Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production[J]. J Appl Phycol, 2009, 21(5): 493-507.
doi: 10.1007/s10811-008-9392-7 URL |
| [33] |
Solovchenko AE. Recent breakthroughs in the biology of astaxanthin accumulation by microalgal cell[J]. Photosynth Res, 2015, 125(3): 437-449.
doi: 10.1007/s11120-015-0156-3 URL |
| [34] |
Mascia F, Girolomoni L, Alcocer MJP, et al. Functional analysis of photosynthetic pigment binding complexes in the green alga Haematococcus pluvialis reveals distribution of astaxanthin in Photosystems[J]. Sci Rep, 2017, 7(1): 16319.
doi: 10.1038/s41598-017-16641-6 URL |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [5] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [6] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [7] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [8] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [9] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [10] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [11] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [12] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [13] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [14] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [15] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||