








生物技术通报 ›› 2021, Vol. 37 ›› Issue (12): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0184
• 研究报告 • 下一篇
詹冬梅1( ), 朱晨1,2, 周承哲1,2, 黄雪婷1, 赖钟雄1,2, 郭玉琼1,2(
), 朱晨1,2, 周承哲1,2, 黄雪婷1, 赖钟雄1,2, 郭玉琼1,2( )
)
收稿日期:2021-02-14
出版日期:2021-12-26
发布日期:2022-01-19
作者简介:詹冬梅,女,硕士研究生,研究方向:茶树栽培育种与生物技术;E-mail: 基金资助:
ZHAN Dong-mei1( ), ZHU Chen1,2, ZHOU Cheng-zhe1,2, HUANG Xue-ting1, LAI Zhong-xiong1,2, GUO Yu-qiong1,2(
), ZHU Chen1,2, ZHOU Cheng-zhe1,2, HUANG Xue-ting1, LAI Zhong-xiong1,2, GUO Yu-qiong1,2( )
)
Received:2021-02-14
Published:2021-12-26
Online:2022-01-19
摘要:
明确Groucho/Thymidine uptake 1(Gro/Tup1)辅转录调控因子在茶树基因组中的数量、结构及其在非生物胁迫和外源激素处理下表达差异,为揭示Gro/Tup1在茶树激素信号转导途径及其在茶树逆境胁迫中的作用奠定理论基础。对茶树Gro/Tup1家族进行鉴定、分析,同时对其编码蛋白理化性质、进化关系、顺式作用元件及其在干旱、低温(4℃)、赤霉素(GA3)、脱落酸(ABA)和茉莉酸甲酯(MeJA)处理下表达模式进行分析。结果显示:茶树Gro/Tup1家族具有13个成员,分为TOPLESS/TOPLESS-related(TPL/TPR)和LEUNIG/LEUNIG HOMOLOG(LUG/LUH)两个亚家族。顺式作用元件预测结果表明,茶树Gro/Tup1家族成员启动子区域有大量与GA、ABA、MeJA响应以及干旱和低温应答的顺式作用元件;RT-qPCR分析结果表明,不同处理下,茶树Gro/Tup1家族成员呈现不同表达模式。重度干旱时,CsLUG3和CsLUG4基因的表达量显著上调,CsTPL2表达水平在轻度干旱时就开始受到抑制,CsTPL8和CsTPL9不受干旱调控;低温处理下CsTPL2和CsTPL3表达量显著上调;GA3处理下,多数基因表达量上调;ABA处理下多数基因表达量均呈现先显著增加后显著下降趋势;MeJA处理下,大部分基因表达水平都受到显著抑制。茶树含有13个Gro/ Tup1家族成员,其结构进化高度保守,能够响应非生物胁迫及外源激素,其基因应答呈现不同表达谱。
詹冬梅, 朱晨, 周承哲, 黄雪婷, 赖钟雄, 郭玉琼. 茶树Gro/Tup1基因家族鉴定及外源激素和非生物胁迫下表达分析[J]. 生物技术通报, 2021, 37(12): 1-12.
ZHAN Dong-mei, ZHU Chen, ZHOU Cheng-zhe, HUANG Xue-ting, LAI Zhong-xiong, GUO Yu-qiong. Genome-wide Identification of Gro/Tup1 Gene Family in Camellia sinensis and Their Expression Analysis Under Exogenous Phytohormones and Abiotic Stress Treatments[J]. Biotechnology Bulletin, 2021, 37(12): 1-12.
| 基因名称 Gene name | 上游引物 Upstream primer | 下游引物 Downstream primer |
|---|---|---|
| CsLUG1 | GCGAGTCATCTGGGTAGTAAGT | GTTGTTGGTGCTCTCTTGCT |
| CsLUG2 | GCTGAGAGTGCTTTGGATGG | GCTGAAGGATGTTGAGTGCT |
| CsLUG3 | TTGGAATGCGAGTGGTCAGT | TGGTGCTGTGGTGTCAACAT |
| CsLUG4 | CGTGGCAAGCCCTTAAAG | TGGTGTCAGCATCTGGAGT |
| CsTPL1 | CATGTGTTGAAGAGACTGAGG | TGGGCAAATCATCAGAAGAG |
| CsTPL2 | GCTACCGCTGTTGAGATTCTTG | ATTTGCCAGGGTGAGGAGTAG |
| CsTPL3 | CCTTCCAACCTACACCAGC | CACCAAGACCAATAGCTCCAC |
| CsTPL4 | TCGGCTTTGTCAGTTCTACG | CCGGAAGTGTTATAGGAGGTG |
| CsTPL5 | GCGACTTGAGCATCTCCTTG | GCTGCCACTTCAAAGCCT |
| CsTPL6 | TGGCTCGCTCTCCATGTATG | TGCAAGAGGAACGTGATGGT |
| CsTPL7 | AGATGTTGGTTTGGTTCAGGG | GCTTTGGTTAATGAGGTTCCG |
| CsTPL8 | TTCAAGGTTGCGGACTCTCAT | GATTGTTTGCTGGTGATGGAG |
| CsTPL9 | GTCGCCTGTAACTAACCCACT | TGAGGAACCGGAGATGGATTAG |
| Csβ-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsSAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
表1 RT-qPCR所用引物序列
Table 1 Primer sequences used in RT-qPCR
| 基因名称 Gene name | 上游引物 Upstream primer | 下游引物 Downstream primer |
|---|---|---|
| CsLUG1 | GCGAGTCATCTGGGTAGTAAGT | GTTGTTGGTGCTCTCTTGCT |
| CsLUG2 | GCTGAGAGTGCTTTGGATGG | GCTGAAGGATGTTGAGTGCT |
| CsLUG3 | TTGGAATGCGAGTGGTCAGT | TGGTGCTGTGGTGTCAACAT |
| CsLUG4 | CGTGGCAAGCCCTTAAAG | TGGTGTCAGCATCTGGAGT |
| CsTPL1 | CATGTGTTGAAGAGACTGAGG | TGGGCAAATCATCAGAAGAG |
| CsTPL2 | GCTACCGCTGTTGAGATTCTTG | ATTTGCCAGGGTGAGGAGTAG |
| CsTPL3 | CCTTCCAACCTACACCAGC | CACCAAGACCAATAGCTCCAC |
| CsTPL4 | TCGGCTTTGTCAGTTCTACG | CCGGAAGTGTTATAGGAGGTG |
| CsTPL5 | GCGACTTGAGCATCTCCTTG | GCTGCCACTTCAAAGCCT |
| CsTPL6 | TGGCTCGCTCTCCATGTATG | TGCAAGAGGAACGTGATGGT |
| CsTPL7 | AGATGTTGGTTTGGTTCAGGG | GCTTTGGTTAATGAGGTTCCG |
| CsTPL8 | TTCAAGGTTGCGGACTCTCAT | GATTGTTTGCTGGTGATGGAG |
| CsTPL9 | GTCGCCTGTAACTAACCCACT | TGAGGAACCGGAGATGGATTAG |
| Csβ-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsSAND | CCAATTGCCCCCTTAATGAC | GCAATCATTTCCTTCGTGGAG |
| 基因名称 Gene name | 基因ID Gene ID | 大小 Size/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index | 脂溶指 Aliphatic index | 亲水指数 GRAVY | 外显子数 Number of exons |
|---|---|---|---|---|---|---|---|---|---|
| CsLUG1 | TEA022439.1 | 864 | 94.51266 | 7.02 | 质体Plastid | 46.11 | 77.78 | 0.342 | 17 |
| CsLUG2 | TEA004970.1 | 660 | 73.86218 | 8.04 | 细胞核Nucleus | 45.09 | 67.7 | 0.566 | 16 |
| CsLUG3 | TEA020168.1 | 907 | 100.83071 | 6.48 | 细胞核Nucleus | 53.22 | 67.42 | 0.648 | 19 |
| CsLUG4 | TEA025268.1 | 757 | 81.54457 | 6.08 | 细胞核Nucleus | 40.86 | 40.86 | 0.352 | 17 |
| CsTPL1 | TEA031994.1 | 540 | 60.18248 | 8.07 | 细胞核Nucleus | 37.53 | 79.65 | 0.368 | 14 |
| CsTPL2 | TEA019471.1 | 1196 | 131.4561 | 6.33 | 细胞核Nucleus | 40.35 | 83.09 | 0.27 | 27 |
| CsTPL3 | TEA016267.1 | 1319 | 145.85055 | 6.87 | 细胞核Nucleus | 39.33 | 77.87 | 0.317 | 30 |
| CsTPL4 | TEA030187.1 | 569 | 64.03845 | 6.27 | 叶绿体Chloroplast | 39.24 | 91.92 | 0.193 | 6 |
| CsTPL5 | TEA024264.1 | 723 | 81.24096 | 8.17 | 细胞核Nucleus | 43.53 | 85.01 | 0.244 | 20 |
| CsTPL6 | TEA022067.1 | 1069 | 118.97318 | 6.14 | 内质网Reticulum | 39.47 | 86.43 | 0.222 | 24 |
| CsTPL7 | TEA022077.1 | 759 | 86.17475 | 6.72 | 细胞核Nucleus | 43.25 | 85.03 | 0.181 | 18 |
| CsTPL8 | TEA020620.1 | 1195 | 131.50275 | 6.43 | 叶绿体Chloroplast | 40.03 | 80.14 | 0.312 | 26 |
| CsTPL9 | TEA023424.1 | 1188 | 131.09747 | 6.3 | 细胞核Nucleus | 38.29 | 79.55 | 0.317 | 26 |
表2 茶树Gro/Tup1蛋白质序列特征
Table 2 Feature of CsGro/Tup1 protein in C. sinensis
| 基因名称 Gene name | 基因ID Gene ID | 大小 Size/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular localization | 不稳定系数 Instability index | 脂溶指 Aliphatic index | 亲水指数 GRAVY | 外显子数 Number of exons |
|---|---|---|---|---|---|---|---|---|---|
| CsLUG1 | TEA022439.1 | 864 | 94.51266 | 7.02 | 质体Plastid | 46.11 | 77.78 | 0.342 | 17 |
| CsLUG2 | TEA004970.1 | 660 | 73.86218 | 8.04 | 细胞核Nucleus | 45.09 | 67.7 | 0.566 | 16 |
| CsLUG3 | TEA020168.1 | 907 | 100.83071 | 6.48 | 细胞核Nucleus | 53.22 | 67.42 | 0.648 | 19 |
| CsLUG4 | TEA025268.1 | 757 | 81.54457 | 6.08 | 细胞核Nucleus | 40.86 | 40.86 | 0.352 | 17 |
| CsTPL1 | TEA031994.1 | 540 | 60.18248 | 8.07 | 细胞核Nucleus | 37.53 | 79.65 | 0.368 | 14 |
| CsTPL2 | TEA019471.1 | 1196 | 131.4561 | 6.33 | 细胞核Nucleus | 40.35 | 83.09 | 0.27 | 27 |
| CsTPL3 | TEA016267.1 | 1319 | 145.85055 | 6.87 | 细胞核Nucleus | 39.33 | 77.87 | 0.317 | 30 |
| CsTPL4 | TEA030187.1 | 569 | 64.03845 | 6.27 | 叶绿体Chloroplast | 39.24 | 91.92 | 0.193 | 6 |
| CsTPL5 | TEA024264.1 | 723 | 81.24096 | 8.17 | 细胞核Nucleus | 43.53 | 85.01 | 0.244 | 20 |
| CsTPL6 | TEA022067.1 | 1069 | 118.97318 | 6.14 | 内质网Reticulum | 39.47 | 86.43 | 0.222 | 24 |
| CsTPL7 | TEA022077.1 | 759 | 86.17475 | 6.72 | 细胞核Nucleus | 43.25 | 85.03 | 0.181 | 18 |
| CsTPL8 | TEA020620.1 | 1195 | 131.50275 | 6.43 | 叶绿体Chloroplast | 40.03 | 80.14 | 0.312 | 26 |
| CsTPL9 | TEA023424.1 | 1188 | 131.09747 | 6.3 | 细胞核Nucleus | 38.29 | 79.55 | 0.317 | 26 |

图2 茶树、拟南芥、番茄和水稻Gro/Tup1蛋白的进化关系与分类
Fig. 2 Phylogenetic relationships and classification of Gro/Tup1 proteins from C. sinensis,A. thaliana,S. lycopersicum,and O. sativa

图3 茶树和拟南芥的Gro/Tup1 A:Gro/Tup1蛋白序列进化关系;B:Gro/Tup1保守基序分布;C:Gro/Tup1基因内含子-外显子结构分布图
Fig. 3 Gro/Tup1 in C. sinensis and A. thaliana A:Sequence evolution relationship of Gro/Tup1 protein. B:Conservative motif distribution of Gro/Tup1. C:Gro/Tup1 gene intron-exon structure distribution
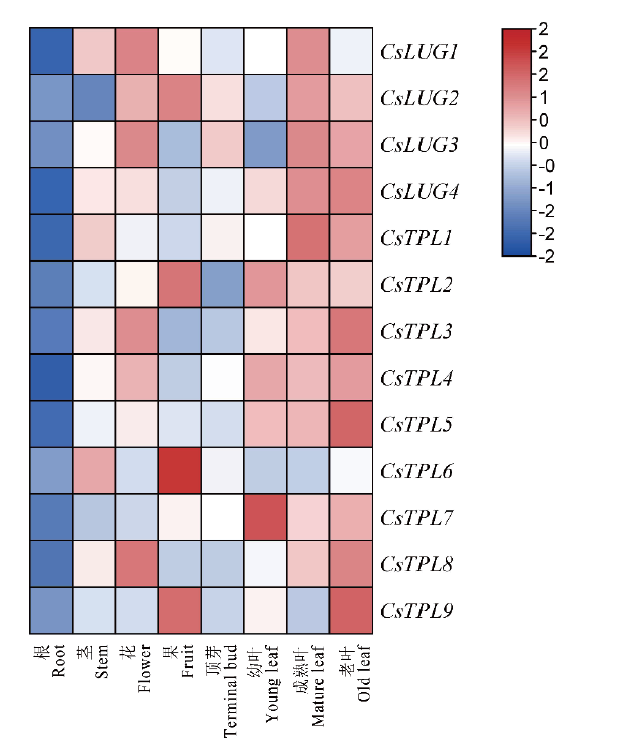
图6 Gro/Tup1家族成员在‘铁观音’组织部位的表达谱 颜色刻度表示log2转换值,红色代表高表达,蓝色代表低表达
Fig.6 Expression profiles of CsGro/Tup1 family genes in the tissues of ‘Tieguanyin’ cultivar The color scale is the log2 converted value. Red indicates high expression,and blue indicates low expression
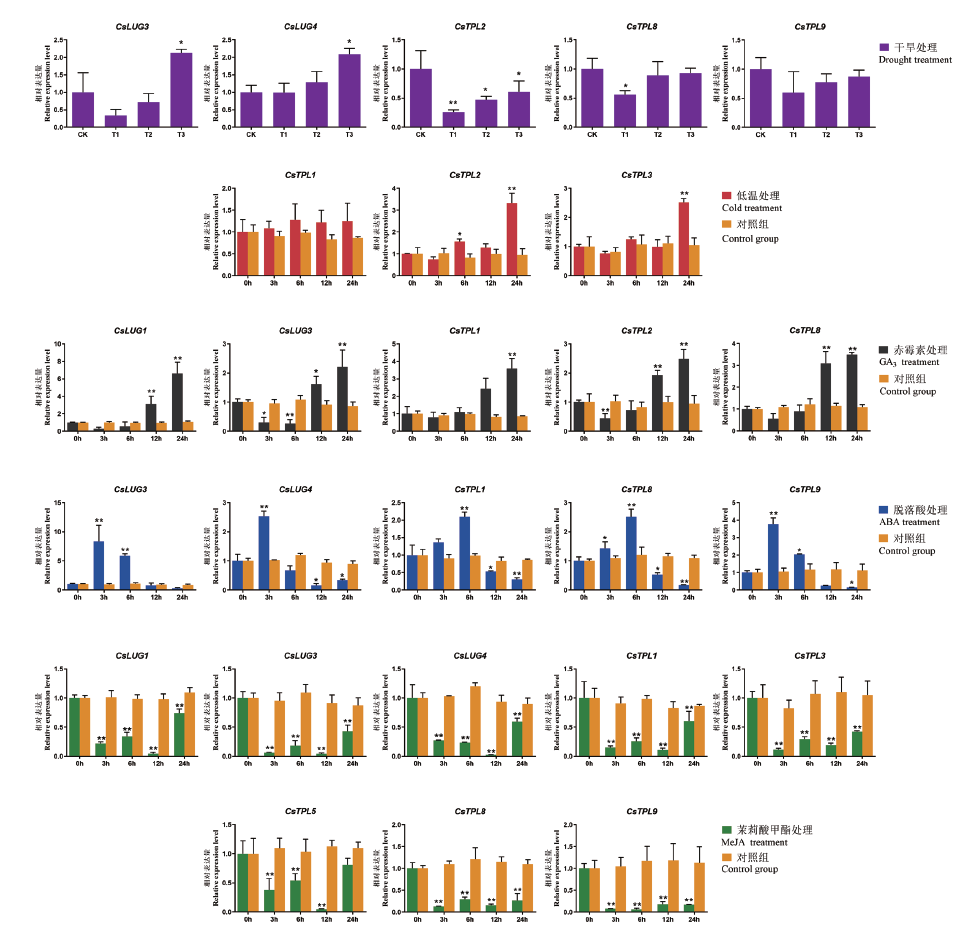
图7 干旱、低温、GA3、ABA和MeJA处理下茶树Gro/Tup1相对表达量
Fig. 7 Expression pattens of CsGro/Tup1 in C. sinensis under drought,cold,GA3,ABA,MeJA treatment *P<0.05,**P<0.01
| [1] |
Courey AJ, Jia S. Transcriptional repression:the long and the short of it[J]. Genes Dev, 2001, 15(21):2786-2796.
doi: 10.1101/gad.939601 URL |
| [2] |
Krogan NT, Long JA. Why so repressed? Turning off transcription during plant growth and development[J]. Curr Opin Plant Biol, 2009, 12(5):628-636.
doi: 10.1016/j.pbi.2009.07.011 pmid: 19700365 |
| [3] |
Mannervik M, Nibu Y, Zhang H, et al. Transcriptional coregulators in development[J]. Science, 1999, 284(5414):606-609.
pmid: 10213677 |
| [4] |
Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression[J]. Gene, 2000, 249(1/2):1-16.
doi: 10.1016/S0378-1119(00)00161-X URL |
| [5] |
Conner J, Liu Z. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development[J]. PNAS, 2000, 97(23):12902-12907.
pmid: 11058164 |
| [6] |
Long JA, Ohno C, Smith ZR, et al. TOPLESS regulates apical embryonic fate in Arabidopsis[J]. Science, 2006, 312(5779):1520-1523.
doi: 10.1126/science.1123841 URL |
| [7] |
Kieffer M, Stern Y, Cook H, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance[J]. Plant Cell, 2006, 18(3):560-573.
pmid: 16461579 |
| [8] |
Liu ZC, Karmarkar V. Groucho/Tup1 family co-repressors in plant development[J]. Trends Plant Sci, 2008, 13(3):137-144.
doi: 10.1016/j.tplants.2007.12.005 URL |
| [9] |
Hao YW, Wang XY, Li X, et al. Genome-wide identification, phylogenetic analysis, expression profiling, and protein-protein interaction properties of TOPLESS gene family members in tomato[J]. J Exp Bot, 2014, 65(4):1013-1023.
doi: 10.1093/jxb/ert440 URL |
| [10] |
Li HY, Huang KF, Du HM, et al. Genome-wide analysis of Gro/Tup1 family corepressors and their responses to hormones and abiotic stresses in maize[J]. J Plant Biol, 2016, 59(6):603-615.
doi: 10.1007/s12374-016-0333-8 URL |
| [11] |
Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis[J]. Science, 2008, 319(5868):1384-1386.
doi: 10.1126/science.1151461 pmid: 18258861 |
| [12] |
Nguyen D, Rieu I, Mariani C, et al. How plants handle multiple stresses:hormonal interactions underlying responses to abiotic stress and insect herbivory[J]. Plant Mol Biol, 2016, 91(6):727-740.
doi: 10.1007/s11103-016-0481-8 pmid: 27095445 |
| [13] |
Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling[J]. Nature, 2010, 464(7289):788-791.
doi: 10.1038/nature08854 URL |
| [14] |
You Y, Zhai Q, An C, et al. LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25[J]. Plant Cell, 2019, 31(9):2187-2205.
doi: 10.1105/tpc.19.00115 URL |
| [15] |
Causier B, Ashworth M, Guo W, et al. The TOPLESS interactome:a framework for gene repression in Arabidopsis[J]. Plant Physiol, 2012, 158(1):423-438.
doi: 10.1104/pp.111.186999 pmid: 22065421 |
| [16] |
Gonzalez D, Bowen AJ, Carroll TS, et al. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14(SWP)and CDK8(HEN3)to repress transcription[J]. Mol Cell Biol, 2007, 27(15):5306-5315.
doi: 10.1128/MCB.01912-06 URL |
| [17] |
Zhu J, Jeong JC, Zhu Y, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance[J]. PNAS, 2008, 105(12):4945-4950.
doi: 10.1073/pnas.0801029105 URL |
| [18] |
Zhu Z, Xu F, Zhang Y, et al. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor[J]. PNAS, 2010, 107(31):13960-13965.
doi: 10.1073/pnas.1002828107 URL |
| [19] |
Shrestha B, Guragain B, Sridhar VV. Involvement of co-repressor LUH and the adapter proteins SLK1 and SLK2 in the regulation of abiotic stress response genes in Arabidopsis[J]. BMC Plant Biol, 2014, 14:54.
doi: 10.1186/1471-2229-14-54 pmid: 24564815 |
| [20] | 芦梅. 中国茶叶产业发展的管理模式与经济学分析[J]. 福建茶叶, 2016, 38(9):99-100. |
| Lu M. Management mode and economic analysis of tea industry development in China[J]. Tea Fujian, 2016, 38(9):99-100. | |
| [21] |
Liu SC, Jin JQ, Ma JQ, et al. Transcriptomic analysis of tea plant responding to drought stress and recovery[J]. PLoS One, 2016, 11(1):e0147306.
doi: 10.1371/journal.pone.0147306 URL |
| [22] |
Cheruiyot EK, Mumera LM, Ng’Etich WK, et al. High fertilizer rates increase susceptibility of tea to water stress[J]. J Plant Nutr, 2009, 33(1):115-129.
doi: 10.1080/01904160903392659 URL |
| [23] |
Wei CL, Yang H, Wang SB, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality[J]. Proc Natl Acad Sci USA, 2018, 115(18):E4151-E4158.
doi: 10.1073/pnas.1719622115 URL |
| [24] |
Xia EH, Zhang HB, Sheng J, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynjournal[J]. Mol Plant, 2017, 10(6):866-877.
doi: 10.1016/j.molp.2017.04.002 URL |
| [25] |
Xia EH, Tong W, Hou Y, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation[J]. Mol Plant, 2020, 13(7):1013-1026.
doi: 10.1016/j.molp.2020.04.010 URL |
| [26] |
Wang X, Feng H, Chang Y, et al. Population sequencing enhances understanding of tea plant evolution[J]. Nat Commun, 2020, 11(1):4447.
doi: 10.1038/s41467-020-18228-8 URL |
| [27] |
Zhang QJ, Li W, Li K, et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution[J]. Mol Plant, 2020, 13(7):935-938.
doi: 10.1016/j.molp.2020.04.009 URL |
| [28] | 项丽慧, 陈林, 余文权, 等. 茶树GH1基因家族鉴定及其在茶鲜叶萎凋过程的表达[J]. 应用与环境生物学报, 2020, 26(4):878-885. |
| Xiang LH, Chen L, Yu WQ, et al. Identification of the GH1 gene family in Camellia sinensis and expression analysis during the withering process of fresh tea leaves[J]. Chin J Appl Environ Biol, 2020, 26(4):878-885. | |
| [29] | 叶一隽, 李佳敏, 曹红利, 等. 茶树CsBBX基因家族的鉴定与表达[J]. 应用与环境生物学报, 2020, 26(6):1508-1516. |
| Ye YJ, Li JM, Cao HL, et al. Identification and expression analysis of the CsBBX gene family in tea plants[J]. Chin J Appl Environ Biol, 2020, 26(6):1508-1516. | |
| [30] |
Xia EH, Li FD, Tong W, et al. Tea Plant Information Archive:a comprehensive genomics and bioinformatics platform for tea plant[J]. Plant Biotechnol J, 2019, 17(10):1938-1953.
doi: 10.1111/pbi.v17.10 URL |
| [31] |
Zhou CZ, Zhu C, Fu H, et al. Genome-wide investigation of superoxide dismutase(SOD)gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant(Camellia sinensis)[J]. PLoS One, 2019, 14(10):e0223609.
doi: 10.1371/journal.pone.0223609 URL |
| [32] |
Ouyang S, Zhu W, Hamilton J, et al. The TIGR Rice Genome Annotation Resource:improvements and new features[J]. Nucleic Acids Res, 2007, 35(database issue):D883-D887.
doi: 10.1093/nar/gkl976 URL |
| [33] | 马洪磊. 植物辅抑制因子TPL/TPR蛋白结构与EAR基序相互作用分子机理研究[D]. 上海:中国科学院上海药物研究所, 2016. |
| Ma HL. Structure of plant co-repressor TPL/TPR protein provides insights into mechanism of EAR motif binding[D]. Shanghai:Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 2016. | |
| [34] |
Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation[J]. Nat Rev Immunol, 2010, 10(1):24-35.
doi: 10.1038/nri2685 pmid: 20029446 |
| [35] |
Stahle MI, Kuehlich J, Staron L, et al. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis[J]. Plant Cell, 2009, 21(10):3105-3118.
doi: 10.1105/tpc.109.070458 URL |
| [36] |
Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1)complex during jasmonate signalling[J]. Nature, 2007, 448(7154):661-665.
doi: 10.1038/nature05960 URL |
| [37] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154):666-671.
doi: 10.1038/nature06006 URL |
| [38] | 彭蕾. DNA序列的Exon-Intron结构的研究[D]. 北京:北京工业大学, 2001. |
| Peng L. Research of exon-intron structures in the DNA sequences[D]. Beijing:Beijing University of Technology, 2001. | |
| [39] | 王晓斌, 刘国仰. 有关内含子功能研究的新进展[J]. 中华医学遗传学杂志, 2000(3):211-212 |
| Wang XB, Liu GY. New research progress on the function of intron[J]. Chin J Med Genet, 2000(3):211-212 | |
| [40] | 单耀军. 内含子与基因间隔序列长度及基因表达量间关系的研究[D]. 保定:河北大学, 2007. |
| Shan YJ. Investigating the relation of introns to the length of intergenic sequences and gene expression levels[D]. Baoding:Hebei University, 2007. | |
| [41] |
Lorenzo O, Chico JM, Sánchez-Serrano JJ, et al. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis[J]. Plant Cell, 2004, 16(7):1938-1950.
pmid: 15208388 |
| [1] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [2] | 刘保财, 陈菁瑛, 张武君, 黄颖桢, 赵云青, 刘剑超, 危智诚. 多花黄精种子微根茎基因表达特征分析[J]. 生物技术通报, 2023, 39(8): 220-233. |
| [3] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [4] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [5] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [6] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [7] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [8] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [9] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [10] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [11] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [12] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [13] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [14] | 于世霞, 姜雨彤, 林文慧. 胚珠原基起始的信号与分子机制研究进展[J]. 生物技术通报, 2023, 39(2): 1-9. |
| [15] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||