








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 97-105.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1344
• 作物品质遗传与改良专题(专题主编: 刘巧泉 教授) • 上一篇 下一篇
Olalekan Amoo( ), 胡利民, 翟云孤, 范楚川(
), 胡利民, 翟云孤, 范楚川( ), 周永明
), 周永明
收稿日期:2021-10-25
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
范楚川,男,博士,教授,研究方向:油菜产量性状遗传解析和基因编辑技术在油菜中的应用;E-mail: fanchuchuan@mail.hzau.edu.cn作者简介:Olalekan Amoo,男,硕士研究生,研究方向:油菜遗传育种;E-mail: olalekan@webmail.hzau.edu.cn基金资助:
Olalekan Amoo( ), HU Li-min, ZHAI Yun-gu, FAN Chu-chuan(
), HU Li-min, ZHAI Yun-gu, FAN Chu-chuan( ), ZHOU Yong-ming
), ZHOU Yong-ming
Received:2021-10-25
Published:2022-04-26
Online:2022-05-06
摘要:
植物的株型决定于其分枝方式,而改良分枝方式是提高作物产量的有效方式之一。目前关于油菜分枝调控的分子机理研究较少,缺乏用于株型改良的种质资源。本研究以油菜BRANCHED1(BRC1)为目的基因,利用CRISPR/Cas9基因编辑技术靶向突变油菜BRC1基因,通过农杆菌介导的遗传转化获得再生苗,PCR技术筛选出转基因阳性单株,再利用Hi-TOM方法进行靶点突变基因型的测序分析,证实基因编辑产生的定点突变可稳定遗传。最后对获得的突变体进行表型观察发现,BRC1的BnaC01g34090D和BnaA01g26700D的双拷贝纯合突变体表现出明显的多分枝表型,研究结果表明BnaBRC1参与油菜中的分枝和株型调控,为后续研究油菜的株型调控分子机理提供了重要研究材料。
Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105.
Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology[J]. Biotechnology Bulletin, 2022, 38(4): 97-105.
| 引物 Primer | 序列 Sequence(5'-3') | 用途 Purpose |
|---|---|---|
| CBnaC01-F1 | GAGACCATAACCACACACAAC | Cloning BnaC01g34090D |
| CBnaC01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaC01-R | TCCATGTAGACAGAGACGATTT | |
| CBnaA01-F1 | GAGACCATAACCACACACAAC | Cloning BnaA01g26700D |
| CBnaA01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaA01-R | GACGATTGCCCACAAAGTAAA | |
| CBnaC05-F | CAGTACTACCACCACCATCAAT | Cloning BnaC05g51920D |
| CBnaC05-R | TCGGGATCTGAGATAGCCTATAA | |
| CBnaCnn-F | ACAACAACAACAGAAGATCTCTCAG | Cloning BnaCnng23770D |
| CBnaCnn-R | CATGAGGTCTCTTGGTCTCTCC | |
| CBnaA03-F1 | ACAACAGGTCTTTCAGTACTACC | Cloning BnaA03g34820D |
| CBnaA03-F2 | CAAGGACCGGCGTTAGG | |
| CBnaA03-R | CATGAGGTCTCTTGGTCTCTC | |
| BnBRC1T1-F | gtcAAGCATCATGCAGGAACATG | BnaBRC1 target construction |
| BnBRC1T1-R | aaacCATGTTCCTGCATGATGCT | |
| BnBRC1T2-F | attGCAAAGGGTCAGGTCCTGAA | |
| BnBRC1T2-R | aaacTTCAGGACCTGACCCTTTG | |
| BnBRC1T3-F | gtcACGGAGAGATTCTCACTCCCA | |
| BnBRC1T3-R | aaacTGGGAGTGAGAATCTCTCCG | |
| BnBRC1T4-F | gtcATGGATATCTCCGTAACTCTC | |
| BnBRC1T4-R | aaacGAGAGTTACGGAGATATCCA | |
| BnBRC1T5-F | attGTCGCCATGTCGAGGACCTCT | |
| BnBRC1T5-R | aaacAGAGGTCCTCGACATGGCGA | |
| BnBRC1T6-F | attGAGGAGATGGTCCATGTCAG | |
| BnBRC1T6-R | aaacCTGACATGGACCATCTCCT | |
| PB-L | GCGCGCgGTctcGCTCGACTAGTATGG | Transgenic positive detection |
| PB-R | GCGCGCggtctcTACCGACGCGTATCC | |
| PSH 20A-1F | ggagtgagtacggtgtgcGAGACCATAACCACACACAAC | BnaBRC1 editing check |
| PSH 20A-1R | gagttggatgctggatggCTCATTCTACGATCTCTTGTCCC | |
| PSH 20A-2F | ggagtgagtacggtgtgcGGGACAAGAGATCGTAGAATGAG | |
| PSH 20A-2R | gagttggatgctggatggATGTATACGTACGGGTTTGGG | |
| PSH 20A-3F | ggagtgagtacggtgtgcACACACATAGAAGATTCCCAGAG | |
| PSH 20A-3R | gagttggatgctggatggAGGCTGTTCGCGATCTTTAT | |
| PSH 20A-4F | ggagtgagtacggtgtgcCCTTTTTCTCACTTCGAATCCG | |
| PSH 20A-4R | gagttggatgctggatggTTGGTTTCTGAGGGCTCAAT | |
| PSH 20B-1F | ggagtgagtacggtgtgcTCTTACAAGCACCTTCTTCTTTTTC | |
| PSH 20B-1R | gagttggatgctggatggCAAGGGCTCAACGAGAGATT | |
| PSH 20B-2F | ggagtgagtacggtgtgcTTGACCACCACCATCATCAG | |
| PSH 20B-2R | gagttggatgctggatggGGGCTCAATGAGGTGAGATT | |
| PSH 20B-CF | ggagtgagtacggtgtgcCGGCACAGCAAGATCAAAAC | |
| PSH 20B-CR | gagttggatgctggatggGCTTGTGTGAGCAACCATTC |
表1 本研究中使用的引物
Table 1 Primers used in this study
| 引物 Primer | 序列 Sequence(5'-3') | 用途 Purpose |
|---|---|---|
| CBnaC01-F1 | GAGACCATAACCACACACAAC | Cloning BnaC01g34090D |
| CBnaC01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaC01-R | TCCATGTAGACAGAGACGATTT | |
| CBnaA01-F1 | GAGACCATAACCACACACAAC | Cloning BnaA01g26700D |
| CBnaA01-F2 | TGTGGACAGTCGAGGATAGA | |
| CBnaA01-R | GACGATTGCCCACAAAGTAAA | |
| CBnaC05-F | CAGTACTACCACCACCATCAAT | Cloning BnaC05g51920D |
| CBnaC05-R | TCGGGATCTGAGATAGCCTATAA | |
| CBnaCnn-F | ACAACAACAACAGAAGATCTCTCAG | Cloning BnaCnng23770D |
| CBnaCnn-R | CATGAGGTCTCTTGGTCTCTCC | |
| CBnaA03-F1 | ACAACAGGTCTTTCAGTACTACC | Cloning BnaA03g34820D |
| CBnaA03-F2 | CAAGGACCGGCGTTAGG | |
| CBnaA03-R | CATGAGGTCTCTTGGTCTCTC | |
| BnBRC1T1-F | gtcAAGCATCATGCAGGAACATG | BnaBRC1 target construction |
| BnBRC1T1-R | aaacCATGTTCCTGCATGATGCT | |
| BnBRC1T2-F | attGCAAAGGGTCAGGTCCTGAA | |
| BnBRC1T2-R | aaacTTCAGGACCTGACCCTTTG | |
| BnBRC1T3-F | gtcACGGAGAGATTCTCACTCCCA | |
| BnBRC1T3-R | aaacTGGGAGTGAGAATCTCTCCG | |
| BnBRC1T4-F | gtcATGGATATCTCCGTAACTCTC | |
| BnBRC1T4-R | aaacGAGAGTTACGGAGATATCCA | |
| BnBRC1T5-F | attGTCGCCATGTCGAGGACCTCT | |
| BnBRC1T5-R | aaacAGAGGTCCTCGACATGGCGA | |
| BnBRC1T6-F | attGAGGAGATGGTCCATGTCAG | |
| BnBRC1T6-R | aaacCTGACATGGACCATCTCCT | |
| PB-L | GCGCGCgGTctcGCTCGACTAGTATGG | Transgenic positive detection |
| PB-R | GCGCGCggtctcTACCGACGCGTATCC | |
| PSH 20A-1F | ggagtgagtacggtgtgcGAGACCATAACCACACACAAC | BnaBRC1 editing check |
| PSH 20A-1R | gagttggatgctggatggCTCATTCTACGATCTCTTGTCCC | |
| PSH 20A-2F | ggagtgagtacggtgtgcGGGACAAGAGATCGTAGAATGAG | |
| PSH 20A-2R | gagttggatgctggatggATGTATACGTACGGGTTTGGG | |
| PSH 20A-3F | ggagtgagtacggtgtgcACACACATAGAAGATTCCCAGAG | |
| PSH 20A-3R | gagttggatgctggatggAGGCTGTTCGCGATCTTTAT | |
| PSH 20A-4F | ggagtgagtacggtgtgcCCTTTTTCTCACTTCGAATCCG | |
| PSH 20A-4R | gagttggatgctggatggTTGGTTTCTGAGGGCTCAAT | |
| PSH 20B-1F | ggagtgagtacggtgtgcTCTTACAAGCACCTTCTTCTTTTTC | |
| PSH 20B-1R | gagttggatgctggatggCAAGGGCTCAACGAGAGATT | |
| PSH 20B-2F | ggagtgagtacggtgtgcTTGACCACCACCATCATCAG | |
| PSH 20B-2R | gagttggatgctggatggGGGCTCAATGAGGTGAGATT | |
| PSH 20B-CF | ggagtgagtacggtgtgcCGGCACAGCAAGATCAAAAC | |
| PSH 20B-CR | gagttggatgctggatggGCTTGTGTGAGCAACCATTC |

图2 BnaBRC1基因的Motif分析及保守结构域分析 A:BRC1同源基因在甘蓝型油菜和拟南芥中的Motif分析;B:油菜和拟南芥中BRC1同源拷贝中保守结构域的预测
Fig.2 Motif analysis and conserved domain structures of BnaBRC1 gene A:Analysis of conserved motifs in the protein sequence of homologous copies of the BRC1 gene in B. napus,and A. thaliana. B:Prediction of the conserved domains in the homologous copies of BRC1 in B. napus and A. thaliana
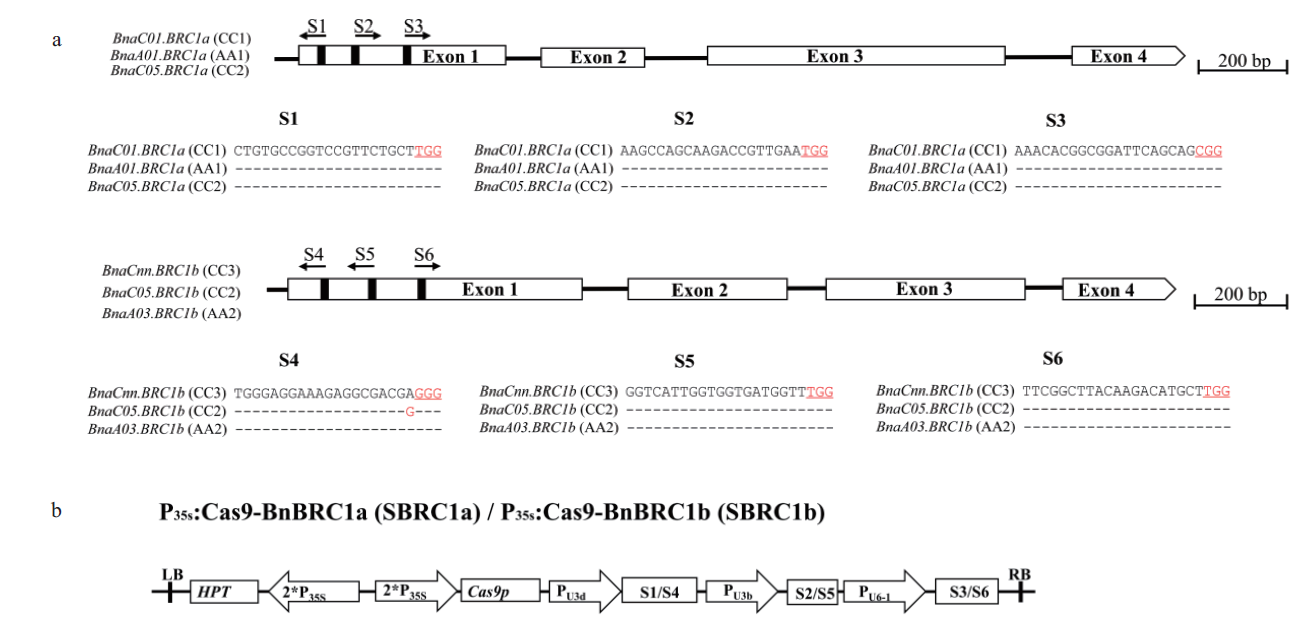
图4 BnaBRC1基因结构与靶点序列及双元质粒载体示意图 a:BnaBRC1基因包括4个外显子,靶点(S1-S6)位置用黑色的竖线标注,方向用带箭头的水平黑线标注。靶点序列中PAM碱基都用红色的字体和下划线强调;b:双元载体SBRC1a和SBRC1b的载体示意图
Fig.4 BnaBRC1 gene structure with target sequences and schematics of binary plasmid vectors a:The BnaBRC1 gene structure includes 4 exons separated by three introns. The vertical line in the gene structure indicates the target site,and the arrow indicates the sgRNA direction. The target sequences are shown with the PAM highlighted in red and underlined. b:Schematic presentation of binary vector SBRC1a and SBRC1b.

图5 BnaBRC1突变体株系在T0代和T1代中S2和S3靶位点突变基因型 CRISPR/Cas9诱导的插入和缺失都用红色的字体或实线表示;PAM序列用红色的字体和下划线强调。“A”,“C”为野生型基因型,“a”,“c”为突变基因型;“-”和“+”分别表示缺失或插入的碱基
Fig.5 Mutated alleles at the S2 and S3 target sites of BnaBRC1 mutants from T0 to T1 generation CRISPR/Cas9-induced insertions and deletions are indicated by red font and red hyphens respectively,while the PAM is underlined and highlighted in red. “A” and “C” are the wild type alleles of the target gene on A and C chromosome copy,and “a”and “c” are the mutated alleles of the target gene on A and C chromosome copy. “_” and “ +” indicate the deletion and insertion of bases,respectively

图6 BnaBRC1突变体的表型观察 A,B:分别为BnaBRC1突变体的叶片数目和株高的统计;C,D:分别为苗期和成熟期野生型材料WT(AA1CC1CC2)和突变体材料(aa1cc1CC2)的株型。其中数据表示平均值+ SD,其中n ≥ 10;标尺=3 cm
Fig.6 Phenotypes of BnaBRC1 mutants A and B:Statistical analysis of leaf numbers and plant height(cm)of BnaBRC1 mutants,respectively. C and D:Phenotypes at the vegetative and flowering stage,respectively,WT refers to wild type(AA1CC1CC2),aacc1CC2 refers to double copy of homozygous BnaBRC1 mutant. The data and error bars represent the mean ± SD,n ≥ 10,and bar = 3 cm
| 靶点 On-target site | 基因 Gene | 测序植株数目 Number of sequenced plants | 脱靶位点数 Putative off-target site | 脱靶编辑 Off-target editing |
|---|---|---|---|---|
| S1 | BnaBRC1 | 72 | 9 | No |
| S2 | BnaBRC1 | 72 | 14 | No |
| S3 | BnaBRC1 | 72 | 11 | No |
| S4 | BnaBRC1 | 54 | 18 | No |
| S5 | BnaBRC1 | 54 | 18 | No |
| S6 | BnaBRC1 | 54 | 16 | No |
表2 在T0代突变体中检测每个sgRNA的脱靶活性
Table 2 Detection of potential off-target effects for each sgRNA target site in T0 mutated plants
| 靶点 On-target site | 基因 Gene | 测序植株数目 Number of sequenced plants | 脱靶位点数 Putative off-target site | 脱靶编辑 Off-target editing |
|---|---|---|---|---|
| S1 | BnaBRC1 | 72 | 9 | No |
| S2 | BnaBRC1 | 72 | 14 | No |
| S3 | BnaBRC1 | 72 | 11 | No |
| S4 | BnaBRC1 | 54 | 18 | No |
| S5 | BnaBRC1 | 54 | 18 | No |
| S6 | BnaBRC1 | 54 | 16 | No |
| [1] |
Jiang HF, Egli DB. Shade induced changes in flower and pod number and flower and fruit abscission in soybean[J]. Agron J, 1993, 85(2):221-225.
doi: 10.2134/agronj1993.00021962008500020011x URL |
| [2] |
Richards RA. Selectable traits to increase crop photosynjournal and yield of grain crops[J]. J Exp Bot, 2000, 51 Spec No:447-458.
doi: 10.1093/jexbot/51.suppl_1.447 URL |
| [3] |
Zhao DL, Atlin GN, Bastiaans L, et al. Developing selection protocols for weed competitiveness in aerobic rice[J]. Field Crops Res, 2006, 97(2/3):272-285.
doi: 10.1016/j.fcr.2005.10.008 URL |
| [4] |
Simon S, Morel K, Durand E, et al. Aphids at crossroads:when branch architecture alters aphid infestation patterns in the apple tree[J]. Trees, 2012, 26(1):273-282.
doi: 10.1007/s00468-011-0629-8 URL |
| [5] |
Zhao WG, Chao HB, Zhang LN, et al. Integration of QTL mapping and gene fishing techniques to dissect the multi-main stem trait in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2019, 10:1152.
doi: 10.3389/fpls.2019.01152 URL |
| [6] | 漆丽萍. 甘蓝型油菜株型与角果相关性状的QTL分析[D]. 武汉:华中农业大学, 2014. |
| Qi LP. Qtl analysis for the traits associated with plant architecture and silique in Brassica napus L.[D]. Wuhan:Huazhong Agricultural University, 2014. | |
| [7] |
Wang YH, Li JY. Genes controlling plant architecture[J]. Curr Opin Biotechnol, 2006, 17(2):123-129.
doi: 10.1016/j.copbio.2006.02.004 URL |
| [8] |
Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize:evidence for epistasis and the evolution of dominance[J]. Genetics, 1995, 141(1):333-346.
doi: 10.1093/genetics/141.1.333 pmid: 8536981 |
| [9] |
Hubbard L, McSteen P, Doebley J , et al. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte[J]. Genetics, 2002, 162(4):1927-1935.
doi: 10.1093/genetics/162.4.1927 pmid: 12524360 |
| [10] |
Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds[J]. Plant Cell, 2007, 19(2):458-472.
pmid: 17307924 |
| [11] |
Dun EA, Brewer PB, Beveridge CA. Strigolactones:discovery of the elusive shoot branching hormone[J]. Trends Plant Sci, 2009, 14(7):364-372.
doi: 10.1016/j.tplants.2009.04.003 URL |
| [12] |
Leyser O. The control of shoot branching:an example of plant information processing[J]. Plant Cell Environ, 2009, 32(6):694-703.
doi: 10.1111/j.1365-3040.2009.01930.x URL |
| [13] |
Beveridge CA, Kyozuka J. New genes in the strigolactone-related shoot branching pathway[J]. Curr Opin Plant Biol, 2010, 13(1):34-39.
doi: 10.1016/j.pbi.2009.10.003 URL |
| [14] | Rameau C, Bertheloot J, Leduc N, et al. Multiple pathways regulate shoot branching[J]. Front Plant Sci, 2015, 5:741. |
| [15] |
Poza-Carrión C, Aguilar-Martínez JA, Cubas P. Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis[J]. Plant Signal Behav, 2007, 2(6):551-552.
doi: 10.4161/psb.2.6.4811 pmid: 19704556 |
| [16] |
Wang M, le Moigne MA, Bertheloot J, et al. BRANCHED1:a key hub of shoot branching[J]. Front Plant Sci, 2019, 10:76.
doi: 10.3389/fpls.2019.00076 URL |
| [17] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8):1274-1284.
doi: 10.1016/j.molp.2015.04.007 URL |
| [18] | 杨阳. 油菜多室基因的鉴定及多室性状形成的分子调控机理解析[D]. 武汉:华中农业大学, 2019. |
| Yang Y. Identification of multilocular gene in Brassica napus and analysis of molecular regulation mechanism of multilocular traits[J]. Wuhan: Huazhong Agricultural University, 2019. | |
| [19] |
Liu Q, Wang C, Jiao XZ, et al. Hi-TOM:a platform for high-throu-ghput tracking of mutations induced by CRISPR/Cas systems[J]. Sci China Life Sci, 2019, 62(1):1-7.
doi: 10.1007/s11427-018-9402-9 URL |
| [20] |
Russell WL, Russell LB, Kelly EM. Radiation dose rate and mutation frequency[J]. Science, 1958, 128(3338):1546-1550.
doi: 10.1126/science.128.3338.1546 URL |
| [21] |
Sega GA. A review of the genetic effects of ethyl methanesulfonate[J]. Mutat Res, 1984, 134(2/3):113-142.
doi: 10.1016/0165-1110(84)90007-1 URL |
| [22] |
Yang Y, Zhu KY, Li HL, et al. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development[J]. Plant Biotechnol J, 2018, 16(7):1322-1335.
doi: 10.1111/pbi.12872 pmid: 29250878 |
| [23] |
Wang YP, Cheng X, Shan QW, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew[J]. Nat Biotechnol, 2014, 32(9):947-951.
doi: 10.1038/nbt.2969 URL |
| [24] | Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems[J]. Philos Trans Royal Soc Lond Ser B Biol Sci, 2019, 374(1772):20180087. |
| [25] |
Braun N, de Saint Germain A, Pillot JP, et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching[J]. Plant Physiol, 2012, 158(1):225-238.
doi: 10.1104/pp.111.182725 pmid: 22045922 |
| [26] |
Dun EA, de Saint Germain A, Rameau C, et al. Antagonistic action of strigolactone and cytokinin in bud outgrowth control[J]. Plant Physiol, 2012, 158(1):487-498.
doi: 10.1104/pp.111.186783 URL |
| [27] |
Lantzouni O, Klermund C, Schwechheimer C. Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings[J]. Plant J, 2017, 92(5):924-938.
doi: 10.1111/tpj.13729 URL |
| [1] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [2] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [3] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [4] | 刘保财, 陈菁瑛, 张武君, 黄颖桢, 赵云青, 刘剑超, 危智诚. 多花黄精种子微根茎基因表达特征分析[J]. 生物技术通报, 2023, 39(8): 220-233. |
| [5] | 石佳鑫, 刘凯, 朱金洁, 祁显涛, 谢传晓, 刘昌林. 基因编辑技术改良玉米株型增加杂交种产量[J]. 生物技术通报, 2023, 39(8): 62-69. |
| [6] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [7] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [8] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [9] | 蔡梦鲜, 高作敏, 胡利娟, 冯群, 王洪程, 朱斌. 天然甘蓝型油菜C染色体组C1,C2缺体的创建及遗传分析[J]. 生物技术通报, 2023, 39(3): 81-88. |
| [10] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [11] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [12] | 支添添, 周舟, 陈纪鹏, 韩成云. 甘蓝型油菜酪氨酸代谢关键基因FAH的克隆、功能鉴定和表达分析[J]. 生物技术通报, 2023, 39(10): 115-127. |
| [13] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [14] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [15] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||