








生物技术通报 ›› 2023, Vol. 39 ›› Issue (12): 16-32.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0793
收稿日期:2023-08-14
出版日期:2023-12-26
发布日期:2024-01-11
通讯作者:
伍国强,男,博士,教授,博士生导师,研究方向:植物逆境生理与基因工程;E-mail: gqwu@lut.edu.cn作者简介:朱业胜,男,硕士研究生,研究方向:植物逆境生理与基因工程;E-mail: 1428209434@qq.com
基金资助:
ZHU Ye-sheng( ), WU Guo-qiang(
), WU Guo-qiang( ), WEI Ming
), WEI Ming
Received:2023-08-14
Published:2023-12-26
Online:2024-01-11
摘要:
植物通过一系列复杂转运系统调节离子稳态以适应盐渍环境,SOS(salt overly sensitive)信号通路是植物响应非生物胁迫的主要信号途径,主要由质膜Na+/H+逆向转运蛋白SOS1、丝氨酸/苏氨酸类蛋白激酶SOS2和钙感应器SOS3组成。SOS1作为SOS信号通路的主要成员之一,广泛存在于高等植物中,由于早期的进化差异,可能导致不同物种SOS1的结构和理化性质存在一定的特异性。SOS1蛋白为一个同型二聚体,每个单体由跨膜和胞内结构域组成,这为整合来自不同途径的信号和调节Na+转运提供了稳定的对接平台。SOS1基因转录水平受到不同胁迫条件的调控,通过Ca2+信号调控、磷酸化、自抑制和与离子转运体协同调控等机制可抑制或激活SOS1活性。该蛋白具有调控植物昼夜节律和pH以及维持离子稳态等功能,在植物逆境胁迫响应中发挥重要作用。论文对SOS1的结构、功能、调控机制及其维持植物离子稳态平衡作用等方面的研究进展加以综述,并对其未来研究方向进行展望,以期为农作物抗逆性遗传改良提供理论支持和优异基因资源。
朱业胜, 伍国强, 魏明. 质膜Na+/H+逆向转运蛋白SOS1在植物离子稳态平衡中的作用[J]. 生物技术通报, 2023, 39(12): 16-32.
ZHU Ye-sheng, WU Guo-qiang, WEI Ming. Roles of Plasma Membrane Na+/H+ Antiporter SOS1 in Maintaining Ionic Homeostasis of Plants[J]. Biotechnology Bulletin, 2023, 39(12): 16-32.
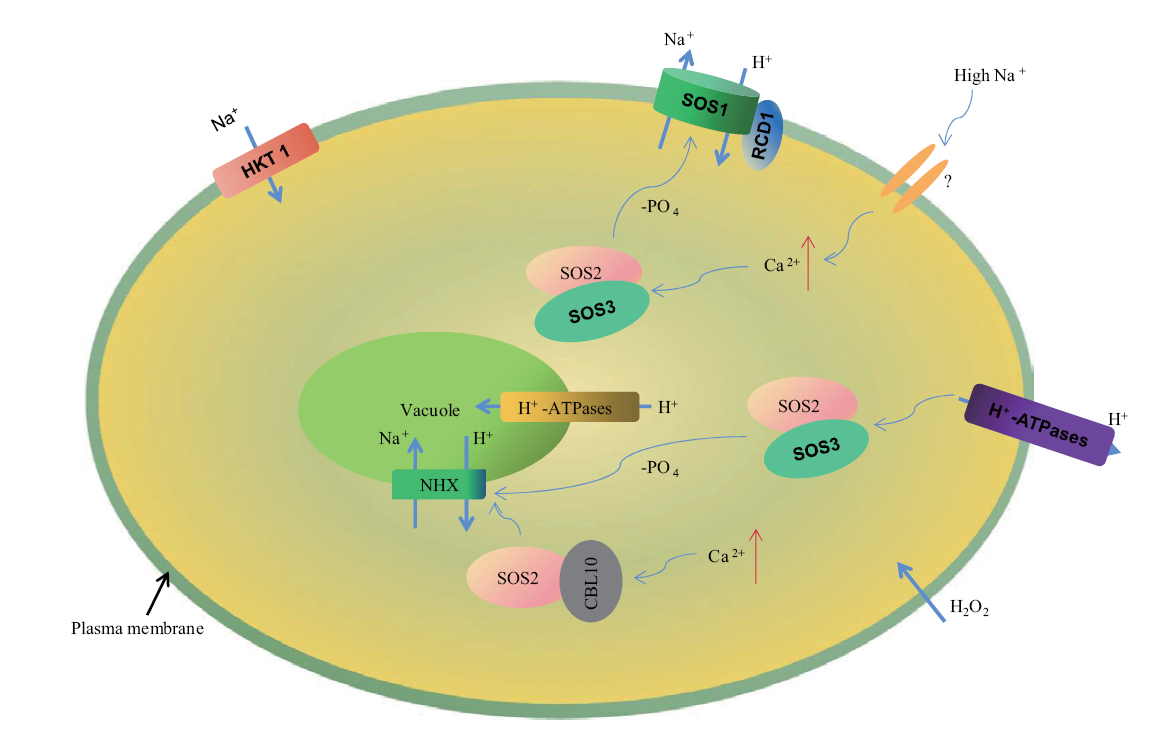
图1 SOS1响应盐胁迫示意图 问号表示不确定性或未知成分。在高盐胁迫下,胞质中Na+水平的升高会导致细胞损伤。细胞中主要的盐解毒机制之一是Ca2+激活的SOS3-SOS2复合物,其激活SOS1并负责Na+外排。同时,SOS3-SOS2复合物也参与HKT1(high-affinity K+ transporter 1)的抑制,HKT1在高盐条件下转运Na+。SOS3家族的另一个成员CBL10也与SOS2形成复合物。SOS3-SOS2复合物可以调节Na+外排(通过调控SOS1)和Na+进入液泡的隔离/区域化(激活NHX并将Na+泵入液泡)。SOS1与RCD1相互作用,减轻活性氧和氧化应激的毒性作用
Fig. 1 Diagrammatic presentation of SOS1 response to salt stress Question marks indicate uncertainties or unknown components. Under high salt stress, the increase of Na+ level in cytoplasm can lead to the damage of multiple cellular processes. One of the main salt detoxification mechanisms in cells is calcium-activated SOS3-SOS2 protein complex, which activates SOS1 and is responsible for the efflux of Na+ out of cells. At the same time, the SOS3-SOS2 complex was also involved in the inhibition of HKT1(high-affinity K+ transporter 1), which transported Na+ under high salt conditions. Another member of the SOS3 family, CBL10, also formed a complex with SOS2. It is hypothesized that this complex can regulate Na+ efflux(by regulating SOS1)and the compartmentation of Na+ into the vacuole(activating NHX and pumping Na+ into the vacuole). SOS1 also interacts with RCD1 and mitigates the toxic effects of ROS and oxidative stress
| 物种名 Species name | 基因名称 Gene name | 基因登录号 GenBank accession No. | 氨基酸 Amino acid | 亚细胞定位 Sub-cellular localization | 跨膜结构域TMD | 亲水性平均值 GRAVY | 等电点 pI | 分子量 Mw/kD | 参考文献 Reference |
|---|---|---|---|---|---|---|---|---|---|
| 甜菜 Beta vulgaris | BvSOS1 | / | 1 162 | PM | 12 | 0.085 | 6.34 | 128.46 | Unpublished |
| 小盐芥thellungiella halophila | ThSOS1 | EF207775 | 1 146 | PM | 11 | 0.090 | 6.40 | 126.35 | [ |
| ThSOS1S | AB562331 | 473 | PM | 11 | 0.722 | 6.15 | 51.19 | [ | |
| 冰叶日中花 Mesembryanthemum crystallinum | McSOS1 | EF207776 | 1 115 | PM | 12 | 0.110 | 5.97 | 127.36 | [ |
| 花花柴 Karelinia caspia | KcSOS1 | MN481368 | 1 136 | PM | 12 | 0.070 | 5.97 | 126.02 | [ |
| 水稻Oryza sativa | OsSOS1-1 | AY785147 | 1 148 | PM | 12 | 0.046 | 6.77 | 127.92 | [ |
| OsSOS1-2 | MG602252 | 1 148 | PM | 13 | 0.047 | 6.77 | 127.90 | ||
| 小麦 Triticum aestivum | TaSOS1-1 | KJ563230 | 1 142 | PM | 12 | 0.110 | 7.00 | 126.22 | [ |
| TaSOS1-2 | AY326952 | 1 142 | PM | 13 | 0.106 | 7.40 | 126.18 | ||
| 大豆 Glycine max | GmSOS1 | JQ287499 | 1 143 | PM | 12 | 0.089 | 6.30 | 126.43 | [ |
| 文冠果Xanthoceras sorbifolium | XsSOS1 | / | 915 | PM | 6 | / | 6.19 | 102.50 | [ |
| 萝卜 Raphanus sativus | RsSOS1 | MZ484950 | 1 137 | PM | 12 | 0.093 | 6.71 | 125.62 | [ |
| 海马齿Sesuvium portulacastrum | SpSOS1 | JX674067 | 1 155 | PM | 12 | 0.063 | 6.08 | 127.95 | [ |
| 印度南瓜 Cucurbita pepo | CmaSOS1 | NW_019272028 | 1 142 | PM | 12 | 0.079 | 5.92 | 126.70 | [ |
| 陆地棉Gossypium hirsutum | GhSOS1 | KU994886 | 1 152 | PM | 12 | 0.061 | 6.32 | 128.18 | [ |
| 拟南芥Arabidopsis thaliana | AtSOS1 | AT2G01980 | 1 146 | PM | 13 | 0.098 | 7.62 | 127.19 | [ |
| 荞麦Fagopyrum esculentum | FeSOS1 | MH064255 | 1 132 | PM | 12 | 0.087 | 6.36 | 126.20 | [ |
| 白刺 Nitraria tangutorum | NtSOS1 | KC292267 | 1 163 | PM | 11 | 0.107 | 6.43 | 127.59 | [ |
| 黄花草木樨Melilotus officinalis | MoSOS1 | MG680455 | 957 | PM | 8 | 0.000 | 6.10 | 107.21 | [ |
| 黄花红砂Reaumuria trigyna | RtSOS1 | KC292265 | 1 145 | PM | 11 | 0.114 | 6.10 | 126.79 | [ |
| 木榄Bruguiera gymnorhiza | BgSOS1 | HM054521 | 1 153 | PM | 12 | 0.136 | 6.19 | 126.91 | [ |
| 盐生草Halogeton glomeratus | HgSOS1 | KT759142 | 1 165 | PM | 11 | 0.086 | 6.21 | 128.97 | / |
| 牡蒿 Artemisia japonica | AjSOS1 | KP896475 | 1 147 | PM | 12 | 0.061 | 6.08 | 127.04 | [ |
| 菠菜 Spinacia oleracea | SoSOS1 | HG799055 | 1 102 | / | 12 | 0.164 | 6.38 | 121.97 | [ |
| 葡萄 Vitis vinifera | VvSOS1 | NM_001281211 | 1 141 | / | 12 | 0.122 | 6.32 | 126.30 | [ |
| 黄瓜 Cucumis sativus | CsSOS1 | JQ655747 | 1 144 | PM | 11 | 0.078 | 6.30 | 127.27 | [ |
| 黑果枸杞Lycium ruthenicum | LrSOS1 | MH454108 | 1 160 | / | 12 | 0.062 | 5.89 | 128.61 | [ |
| 欧洲油菜 Brassica napus | BnSOS1 | EU487184 | 1 142 | PM | 11 | 0.109 | 6.45 | 126.01 | [ |
| 胡杨 Populus euphratica | PeSOS1 | KX132789 | 1 145 | PM | 12 | 0.141 | 6.43 | 126.84 | [ |
| 篦麻 Ricinus communis | RcSOS1 | KX943304 | 1 143 | PM | 12 | 0.069 | 6.62 | 126.49 | [ |
| 菊芋 Helianthus tuberosus | HtSOS1 | KC410809 | 1 130 | PM | 12 | 0.091 | 6.00 | 125.00 | [ |
| 盐地碱蓬 Suaeda salsa | SsSOS1 | KF914414.1 | 1 169 | PM | 12 | 0.023 | 6.52 | 129.20 | [ |
| 绿豆 Vigna radiata | VrSOS1 | KC855193 | 1 046 | / | 11 | 0.119 | 6.20 | 116.10 | / |
| 节节麦 Aegilops tauschii | AtASOS1 | FN356231 | 1 142 | / | 12 | 0.10 | 6.85 | 126.18 | / |
| 短柄草Brachypodium sylvaticum | BsSOS1 | KF671967 | 1 140 | / | 12 | 0.078 | 7.63 | 126.77 | / |
| 海滨锦葵Kosteletzkya virginica | KvSOS1 | KJ577576 | 1 147 | PM | 12 | 0.073 | 6.18 | 127.56 | [ |
| 山萮菜Eutrema salsugineum | EsSOS1 | KF671959 | 1 127 | / | 12 | 0.064 | 6.47 | 124.51 | [ |
| 一粒小麦Triticum monococcum | TmSOS1 | FN356229 | 1 142 | / | 12 | 0.108 | 6.87 | 126.33 | / |
| 大叶补血草Limonium gmelinii | LgSOS1 | EU780458 | 1 151 | PM | 13 | 0.163 | 6.15 | 126.52 | [ |
| 藜麦Chenopodium quinoa | CqSOS1 | EU024570 | 1 158 | PM | 11 | 0.094 | 6.27 | 127.90 | [ |
| 霸王Zygophyllum xanthoxylon | ZxSOS1 | GU177864 | 1 153 | PM | 12 | 0.083 | 6.20 | 127.88 | [ |
| 番茄Solanum lycopersicum | SlSOS1-1 | AB675690 | 1 151 | PM | 12 | 0.069 | 5.85 | 127.54 | [ |
| SlSOS1-2 | NM_001247769 | 1 151 | PM | 13 | 0.083 | 5.89 | 127.50 | ||
| 苦荞麦Fagopyrum tataricum | FtSOS1 | KY659584 | 1 131 | PM | 12 | 0.090 | 6.13 | 125.88 | [ |
| 盐角草Salicornia brachiata | SbSOS1 | EU879059 | 1 159 | / | 11 | 0.037 | 5.92 | 128.55 | [ |
| 山丹 Lilium pumilum | LpSOS1 | OM650676 | 1 161 | PM | 12 | 0.107 | 6.59 | 129.14 | [ |
| 甘蔗Saccharum officinarum | ScSOS1 | KT003285 | 423 | Nucleus | / | -0.396 | 9.12 | 47.60 | [ |
| 小花碱茅Puccinellia tenuiflora | PtSOS1-1 | EF440291 | 1 137 | PM | 11 | 0.107 | 6.48 | 125.50 | [ |
| PtSOS1-2 | GQ452778 | 1 143 | PM | 13 | 0.103 | 6.82 | 126.16 | ||
| 海岛棉Gossypium barbadense | GbSOS1 | / | 1 152 | PM | / | / | 6.42 | 128.04 | [ |
表1 不同植物SOS1基因
Table 1 SOS1 gene in different plant species
| 物种名 Species name | 基因名称 Gene name | 基因登录号 GenBank accession No. | 氨基酸 Amino acid | 亚细胞定位 Sub-cellular localization | 跨膜结构域TMD | 亲水性平均值 GRAVY | 等电点 pI | 分子量 Mw/kD | 参考文献 Reference |
|---|---|---|---|---|---|---|---|---|---|
| 甜菜 Beta vulgaris | BvSOS1 | / | 1 162 | PM | 12 | 0.085 | 6.34 | 128.46 | Unpublished |
| 小盐芥thellungiella halophila | ThSOS1 | EF207775 | 1 146 | PM | 11 | 0.090 | 6.40 | 126.35 | [ |
| ThSOS1S | AB562331 | 473 | PM | 11 | 0.722 | 6.15 | 51.19 | [ | |
| 冰叶日中花 Mesembryanthemum crystallinum | McSOS1 | EF207776 | 1 115 | PM | 12 | 0.110 | 5.97 | 127.36 | [ |
| 花花柴 Karelinia caspia | KcSOS1 | MN481368 | 1 136 | PM | 12 | 0.070 | 5.97 | 126.02 | [ |
| 水稻Oryza sativa | OsSOS1-1 | AY785147 | 1 148 | PM | 12 | 0.046 | 6.77 | 127.92 | [ |
| OsSOS1-2 | MG602252 | 1 148 | PM | 13 | 0.047 | 6.77 | 127.90 | ||
| 小麦 Triticum aestivum | TaSOS1-1 | KJ563230 | 1 142 | PM | 12 | 0.110 | 7.00 | 126.22 | [ |
| TaSOS1-2 | AY326952 | 1 142 | PM | 13 | 0.106 | 7.40 | 126.18 | ||
| 大豆 Glycine max | GmSOS1 | JQ287499 | 1 143 | PM | 12 | 0.089 | 6.30 | 126.43 | [ |
| 文冠果Xanthoceras sorbifolium | XsSOS1 | / | 915 | PM | 6 | / | 6.19 | 102.50 | [ |
| 萝卜 Raphanus sativus | RsSOS1 | MZ484950 | 1 137 | PM | 12 | 0.093 | 6.71 | 125.62 | [ |
| 海马齿Sesuvium portulacastrum | SpSOS1 | JX674067 | 1 155 | PM | 12 | 0.063 | 6.08 | 127.95 | [ |
| 印度南瓜 Cucurbita pepo | CmaSOS1 | NW_019272028 | 1 142 | PM | 12 | 0.079 | 5.92 | 126.70 | [ |
| 陆地棉Gossypium hirsutum | GhSOS1 | KU994886 | 1 152 | PM | 12 | 0.061 | 6.32 | 128.18 | [ |
| 拟南芥Arabidopsis thaliana | AtSOS1 | AT2G01980 | 1 146 | PM | 13 | 0.098 | 7.62 | 127.19 | [ |
| 荞麦Fagopyrum esculentum | FeSOS1 | MH064255 | 1 132 | PM | 12 | 0.087 | 6.36 | 126.20 | [ |
| 白刺 Nitraria tangutorum | NtSOS1 | KC292267 | 1 163 | PM | 11 | 0.107 | 6.43 | 127.59 | [ |
| 黄花草木樨Melilotus officinalis | MoSOS1 | MG680455 | 957 | PM | 8 | 0.000 | 6.10 | 107.21 | [ |
| 黄花红砂Reaumuria trigyna | RtSOS1 | KC292265 | 1 145 | PM | 11 | 0.114 | 6.10 | 126.79 | [ |
| 木榄Bruguiera gymnorhiza | BgSOS1 | HM054521 | 1 153 | PM | 12 | 0.136 | 6.19 | 126.91 | [ |
| 盐生草Halogeton glomeratus | HgSOS1 | KT759142 | 1 165 | PM | 11 | 0.086 | 6.21 | 128.97 | / |
| 牡蒿 Artemisia japonica | AjSOS1 | KP896475 | 1 147 | PM | 12 | 0.061 | 6.08 | 127.04 | [ |
| 菠菜 Spinacia oleracea | SoSOS1 | HG799055 | 1 102 | / | 12 | 0.164 | 6.38 | 121.97 | [ |
| 葡萄 Vitis vinifera | VvSOS1 | NM_001281211 | 1 141 | / | 12 | 0.122 | 6.32 | 126.30 | [ |
| 黄瓜 Cucumis sativus | CsSOS1 | JQ655747 | 1 144 | PM | 11 | 0.078 | 6.30 | 127.27 | [ |
| 黑果枸杞Lycium ruthenicum | LrSOS1 | MH454108 | 1 160 | / | 12 | 0.062 | 5.89 | 128.61 | [ |
| 欧洲油菜 Brassica napus | BnSOS1 | EU487184 | 1 142 | PM | 11 | 0.109 | 6.45 | 126.01 | [ |
| 胡杨 Populus euphratica | PeSOS1 | KX132789 | 1 145 | PM | 12 | 0.141 | 6.43 | 126.84 | [ |
| 篦麻 Ricinus communis | RcSOS1 | KX943304 | 1 143 | PM | 12 | 0.069 | 6.62 | 126.49 | [ |
| 菊芋 Helianthus tuberosus | HtSOS1 | KC410809 | 1 130 | PM | 12 | 0.091 | 6.00 | 125.00 | [ |
| 盐地碱蓬 Suaeda salsa | SsSOS1 | KF914414.1 | 1 169 | PM | 12 | 0.023 | 6.52 | 129.20 | [ |
| 绿豆 Vigna radiata | VrSOS1 | KC855193 | 1 046 | / | 11 | 0.119 | 6.20 | 116.10 | / |
| 节节麦 Aegilops tauschii | AtASOS1 | FN356231 | 1 142 | / | 12 | 0.10 | 6.85 | 126.18 | / |
| 短柄草Brachypodium sylvaticum | BsSOS1 | KF671967 | 1 140 | / | 12 | 0.078 | 7.63 | 126.77 | / |
| 海滨锦葵Kosteletzkya virginica | KvSOS1 | KJ577576 | 1 147 | PM | 12 | 0.073 | 6.18 | 127.56 | [ |
| 山萮菜Eutrema salsugineum | EsSOS1 | KF671959 | 1 127 | / | 12 | 0.064 | 6.47 | 124.51 | [ |
| 一粒小麦Triticum monococcum | TmSOS1 | FN356229 | 1 142 | / | 12 | 0.108 | 6.87 | 126.33 | / |
| 大叶补血草Limonium gmelinii | LgSOS1 | EU780458 | 1 151 | PM | 13 | 0.163 | 6.15 | 126.52 | [ |
| 藜麦Chenopodium quinoa | CqSOS1 | EU024570 | 1 158 | PM | 11 | 0.094 | 6.27 | 127.90 | [ |
| 霸王Zygophyllum xanthoxylon | ZxSOS1 | GU177864 | 1 153 | PM | 12 | 0.083 | 6.20 | 127.88 | [ |
| 番茄Solanum lycopersicum | SlSOS1-1 | AB675690 | 1 151 | PM | 12 | 0.069 | 5.85 | 127.54 | [ |
| SlSOS1-2 | NM_001247769 | 1 151 | PM | 13 | 0.083 | 5.89 | 127.50 | ||
| 苦荞麦Fagopyrum tataricum | FtSOS1 | KY659584 | 1 131 | PM | 12 | 0.090 | 6.13 | 125.88 | [ |
| 盐角草Salicornia brachiata | SbSOS1 | EU879059 | 1 159 | / | 11 | 0.037 | 5.92 | 128.55 | [ |
| 山丹 Lilium pumilum | LpSOS1 | OM650676 | 1 161 | PM | 12 | 0.107 | 6.59 | 129.14 | [ |
| 甘蔗Saccharum officinarum | ScSOS1 | KT003285 | 423 | Nucleus | / | -0.396 | 9.12 | 47.60 | [ |
| 小花碱茅Puccinellia tenuiflora | PtSOS1-1 | EF440291 | 1 137 | PM | 11 | 0.107 | 6.48 | 125.50 | [ |
| PtSOS1-2 | GQ452778 | 1 143 | PM | 13 | 0.103 | 6.82 | 126.16 | ||
| 海岛棉Gossypium barbadense | GbSOS1 | / | 1 152 | PM | / | / | 6.42 | 128.04 | [ |

图2 SOS1 蛋白结构示意图 红色框表示TMD;蓝色箭头表示SOS2-SOS3复合物识别保守磷酸化位点
Fig. 2 Structural diagram of SOS1 protein The red box represents the TMD. The blue arrow indicates the conserved phosphorylation site recognized by SOS2-SOS3 complex

图3 植物SOS1基因家族系统发育树 SOS1基因的来源、名称及蛋白登录号如下:小盐芥ThSOS1(ABN04857);獐茅AlSOS1(AEV89922);拟斯卑尔脱山羊草AsSOS1(CAX83736);白桦BpSOS1(ALV66191);雏菊CcSOS1(BAR88076);海草CnoSOS1(CAD20320);欧亚香花芥HmSOS1(AHL27892);中华大节竹IsSOS1(AGB06353);灰胡杨PpSOS1(AQN76710);碱蓬粳稻SjSOS1(BAE95196);马铃薯StSOS1(XP_006364070);硬粒小麦TtSOS1(ACB47885)。其他物种、基因名称和登录号如表1所示
Fig. 3 Phylogenetic relationship of plant SOS1 gene family The source, name and accession number of the SOS1 family are as follows: Thellungiella salsuginea ThSOS1(ABN04857); Aeluropus littoralis AlSOS1(AEV89922); Aegilops speltoides AsSOS1(CAX83736); Betula platyphylla BpSOS1(ALV66191); Chrysanthemum crissum CcSOS1(BAR88076); Cymodocea nodosa CnoSOS1(CAD20320); Hesperis matronalis HmSOS1(AHL27892); Indosasa sinica IsSOS1(AGB06353); Populus pruinosa PpSOS1(AQN76710); Suaeda japonica SjSOS1(BAE95196); Solanum tuberosum StSOS1(XP_006364070); Triticum turgidum TtSOS1(ACB47885). Other species, name and accession number of the SOS genes are shown in Table 1
| [1] |
Ma L, Liu XH, Lv WJ, et al. Molecular mechanisms of plant responses to salt stress[J]. Front Plant Sci, 2022, 13: 934877.
doi: 10.3389/fpls.2022.934877 URL |
| [2] |
Foster KJ, Miklavcic SJ. A comprehensive biophysical model of ion and water transport in plant roots. II. clarifying the roles of SOS1 in the salt-stress response in Arabidopsis[J]. Front Plant Sci, 2019, 10: 1121.
doi: 10.3389/fpls.2019.01121 pmid: 31620152 |
| [3] |
Zhu JK. Salt and drought stress signal transduction in plants[J]. Annu Rev Plant Biol, 2002, 53: 247-273.
doi: 10.1146/arplant.2002.53.issue-1 URL |
| [4] |
Zhu M, Shabala L, Cuin TA, et al. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat[J]. J Exp Bot, 2016, 67(3): 835-844.
doi: 10.1093/jxb/erv493 URL |
| [5] |
Ishikawa T, Shabala S. Control of xylem Na+ loading and transport to the shoot in rice and barley as a determinant of differential salinity stress tolerance[J]. Physiol Plant, 2019, 165(3): 619-631.
doi: 10.1111/ppl.12758 pmid: 29761494 |
| [6] |
Wu HH, Zhang XC, Giraldo JP, et al. It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress[J]. Plant Soil, 2018, 431(1): 1-17.
doi: 10.1007/s11104-018-3770-y |
| [7] |
Isayenkov SV, Maathuis FJM. Plant salinity stress: many unanswered questions remain[J]. Front Plant Sci, 2019, 10: 80.
doi: 10.3389/fpls.2019.00080 pmid: 30828339 |
| [8] |
Rubio F, Nieves-Cordones M, Horie T, et al. Doing ‘business as usual’ comes with a cost: evaluating energy cost of maintaining plant intracellular K+ homeostasis under saline conditions[J]. New Phytol, 2020, 225(3): 1097-1104.
doi: 10.1111/nph.v225.3 URL |
| [9] |
Wu N, Li Z, Wu F, et al. Sex-specific photosynthetic capacity and Na+ homeostasis in Populus euphratica exposed to NaCl stress and AMF inoculation[J]. Front Plant Sci, 2022, 13: 1066954.
doi: 10.3389/fpls.2022.1066954 URL |
| [10] |
Gul Z, Tang ZH, Arif M, et al. An insight into abiotic stress and influx tolerance mechanisms in plants to cope in saline environments[J]. Biology, 2022, 11(4): 597.
doi: 10.3390/biology11040597 URL |
| [11] |
Gupta BK, Sahoo KK, Anwar K, et al. Silicon nutrition stimulates salt-overly sensitive(SOS)pathway to enhance salinity stress tolerance and yield in rice[J]. Plant Physiol Biochem, 2021, 166: 593-604.
doi: 10.1016/j.plaphy.2021.06.010 URL |
| [12] |
DeTar RA, Höhner R, Manavski N, et al. Loss of salt overly sensitive 1 prevents virescence in chloroplast K+/H+ efflux antiporter-deficient mutants[J]. Plant Physiol, 2022, 189(3): 1220-1225.
doi: 10.1093/plphys/kiac142 URL |
| [13] |
Wang YH, Pan CC, Chen QH, et al. Architecture and autoinhibitory mechanism of the plasma membrane Na+/H+ antiporter SOS1 in Arabidopsis[J]. Nat Commun, 2023, 14(1): 4487.
doi: 10.1038/s41467-023-40215-y |
| [14] |
Ponce KS, Meng LJ, Guo LB, et al. Advances in sensing, response and regulation mechanism of salt tolerance in rice[J]. Int J Mol Sci, 2021, 22(5): 2254.
doi: 10.3390/ijms22052254 URL |
| [15] |
Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition[J]. Plant Cell, 1998, 10(7): 1181-1191.
doi: 10.1105/tpc.10.7.1181 pmid: 9668136 |
| [16] |
Shi H, Ishitani M, Kim C, et al. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter[J]. Proc Natl Acad Sci USA, 2000, 97(12): 6896-6901.
doi: 10.1073/pnas.120170197 pmid: 10823923 |
| [17] |
Wu SJ, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition[J]. Plant Cell, 1996, 8(4): 617-627.
doi: 10.2307/3870339 URL |
| [18] |
Shi HZ, Quintero FJ, Pardo JM, et al. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants[J]. Plant Cell, 2002, 14(2): 465-477.
doi: 10.1105/tpc.010371 URL |
| [19] |
Che BN, Cheng C, Fang JJ, et al. The recretohalophyte Tamarix TrSOS1 gene confers enhanced salt tolerance to transgenic hairy root composite cotton seedlings exhibiting virus-induced gene silencing of GhSOS1[J]. Int J Mol Sci, 2019, 20(12): 2930.
doi: 10.3390/ijms20122930 URL |
| [20] |
时丕彪, 洪立洲, 王军, 等. 印度南瓜Na+/H+逆向转运蛋白基因CmaSOS1的克隆与表达分析[J]. 核农学报, 2020, 34(12): 2638-2646.
doi: 10.11869/j.issn.100-8551.2020.12.2638 |
| Shi PB, Hong LZ, Wang J, et al. Cloning and expression analysis of a Na+/H+antiporter gene CmaSOS1 in Cucurbita maxima[J]. J Nucl Agric Sci, 2020, 34(12): 2638-2646. | |
| [21] |
Shahzad B, Shabala L, Zhou MX, et al. Comparing essentiality of SOS1-mediated Na+ exclusion in salinity tolerance between cultivated and wild rice species[J]. Int J Mol Sci, 2022, 23(17): 9900.
doi: 10.3390/ijms23179900 URL |
| [22] |
El Mahi H, Pérez-Hormaeche J, Luca AD, et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice[J]. Plant Physiol, 2019, 180(2): 1046-1065.
doi: 10.1104/pp.19.00324 URL |
| [23] |
Tomita M, Yamashita M, Omichi A. Gene structure of three kinds of vacuolar-type Na+/H+ antiporters including TaNHX2 transcribed in bread wheat[J]. Genet Mol Biol, 2021, 44(1): e20200207.
doi: 10.1590/1678-4685-gmb-2020-0207 URL |
| [24] |
Jiang W, Pan R, Buitrago S, et al. Conservation and divergence of the TaSOS1 gene family in salt stress response in wheat(Triticum aestivum L.)[J]. Physiol Mol Biol Plants, 2021, 27(6): 1245-1260.
doi: 10.1007/s12298-021-01009-y |
| [25] |
Li SJ, Wu GQ, Lin LY. AKT1, HAK5, SKOR, HKT1;5, SOS1 and NHX1 synergistically control Na+ and K+ homeostasis in sugar beet(Beta vulgaris L.)seedlings under saline conditions[J]. J Plant Biochem Biotechnol, 2022, 31(1): 71-84.
doi: 10.1007/s13562-021-00656-2 |
| [26] |
Zhang WT, Li JX, Dong JH, et al. RsSOS1 responding to salt stress might be involved in regulating salt tolerance by maintaining Na+ homeostasis in radish(Raphanus sativus L.)[J]. Horticulturae, 2021, 7(11): 458.
doi: 10.3390/horticulturae7110458 URL |
| [27] | 张婷婷, 康宇乾, 李雨欣, 等. 非生物胁迫下海马齿SpSOS1基因的表达及响应ABA模式分析[J]. 分子植物育种, 2021, 19(13): 4371-4377. |
| Zhang TT, Kang YQ, Li YX, et al. Expression pattern analysis of SpSOS1 from Sesuvium portulacastrum under abiotic stresses and the response to ABA[J]. Mol Plant Breed, 2021, 19(13): 4371-4377. | |
| [28] |
Zhao CY, William D, Sandhu D. Isolation and characterization of salt overly sensitive family genes in spinach[J]. Physiol Plant, 2021, 171(4): 520-532.
doi: 10.1111/ppl.v171.4 URL |
| [29] |
Brindha C, Vasantha S, Raja AK, et al. Characterization of the salt overly sensitive pathway genes in sugarcane under salinity stress[J]. Physiol Plant, 2021, 171(4): 677-687.
doi: 10.1111/ppl.13245 pmid: 33063359 |
| [30] |
Yang YQ, Han XL, Ma L, et al. Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress[J]. Mol Plant, 2021, 14(12): 2000-2014.
doi: 10.1016/j.molp.2021.07.020 URL |
| [31] |
Xie Q, Zhou Y, Jiang XY. Structure, function, and regulation of the plasma membrane Na+/H+ antiporter salt overly sensitive 1 in plants[J]. Front Plant Sci, 2022, 13: 866265.
doi: 10.3389/fpls.2022.866265 URL |
| [32] |
Zhang MH, Cao JF, Zhang TX, et al. A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max[J]. Front Plant Sci, 2022, 13: 870695.
doi: 10.3389/fpls.2022.870695 URL |
| [33] | 陈叶, 丁健, 阮成江, 等. 文冠果Na+/H+逆向转运蛋白基因XsSOS1的克隆与表达分析[J]. 分子植物育种, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20220803.1806.016.html. |
| Chen Y, Ding J, Ruan CJ, et al. Cloning and expression analysis of a Na+/H+ antiporter gene XsSOS1 in Xanthoceras sorbifolium[J]. Mol Plant Breed, 2022. http://kns.cnki.net/kcms/detail/46.1068.S.20220803.1806.016.html. | |
| [34] |
Arciniegas Vega JP, Melino VJ. Uncovering natural genetic variants of the SOS pathway to improve salinity tolerance in maize[J]. New Phytol, 2022, 236(2): 313-315.
doi: 10.1111/nph.18422 pmid: 35977055 |
| [35] |
Xu FC, Wang MJ, Guo YW, et al. The Na+/H+ antiporter GbSOS1 interacts with SIP5 and regulates salt tolerance in Gossypium barbadense[J]. Plant Sci, 2023, 330: 111658.
doi: 10.1016/j.plantsci.2023.111658 URL |
| [36] |
Liu ZY, Xie QJ, Tang FF, et al. The ThSOS3 gene improves the salt tolerance of transgenic Tamarix hispida and Arabidopsis thaliana[J]. Front Plant Sci, 2021, 11: 597480.
doi: 10.3389/fpls.2020.597480 URL |
| [37] |
Taji T, Komatsu K, Katori T, et al. Comparative genomic analysis of 1047 completely sequenced cDNAs from an Arabidopsis-related model halophyte, Thellungiella halophila[J]. BMC Plant Biol, 2010, 10: 261.
doi: 10.1186/1471-2229-10-261 URL |
| [38] |
Cosentino C, Fischer-Schliebs E, Bertl A, et al. Na+/H+ antiporters are differentially regulated in response to NaCl stress in leaves and roots of Mesembryanthemum crystallinum[J]. New Phytol, 2010, 186(3): 669-680.
doi: 10.1111/j.1469-8137.2010.03208.x pmid: 20298477 |
| [39] |
Guo Q, Meng L, Han JW, et al. SOS1 is a key systemic regulator of salt secretion and K+/Na+ homeostasis in the recretohalophyte Karelinia caspia[J]. Environ Exp Bot, 2020, 177: 104098.
doi: 10.1016/j.envexpbot.2020.104098 URL |
| [40] |
Chen XG, Lu XK, Shu N, et al. GhSOS1, a plasma membrane Na+/H+ antiporter gene from upland cotton, enhances salt tolerance in transgenic Arabidopsis thaliana[J]. PLoS One, 2017, 12(7): e0181450.
doi: 10.1371/journal.pone.0181450 URL |
| [41] |
郑琳琳, 张慧荣, 贺龙梅, 等. 唐古特白刺质膜Na+/H+逆向转运蛋白基因的克隆与表达分析[J]. 草业学报, 2013, 22(4): 179-186.
doi: 10.11686/cyxb20130422 |
| Zheng LL, Zhang HR, He LM, et al. Isolation and expression analysis of a plasma membrane Na+/H+ antiporter from Nitraria tangutorum[J]. Acta Prataculturae Sin, 2013, 22(4): 179-186. | |
| [42] |
黄坤勇, 李杉杉, 郭强, 等. 黄花草木樨MoSOS1基因克隆及表达分析[J]. 生物技术通报, 2017, 33(9): 120-130.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0364 |
| Huang KY, Li SS, Guo Q, et al. Cloning and expression analysis of MoSOS1 gene in Melilotus officinalis[J]. Biotechnol Bull, 2017, 33(9): 120-130. | |
| [43] |
Wang JY, Li Q, Zhang M, et al. The high pH value of alkaline salt destroys the root membrane permeability of Reaumuria trigyna and leads to its serious physiological decline[J]. J Plant Res, 2022, 135(6): 785-798.
doi: 10.1007/s10265-022-01410-y |
| [44] | 荆海瑜, 周扬, 郭晓颖, 等. 木榄细胞膜Na+/H+逆向运输蛋白BgSOS1功能的初步验证[J]. 海南大学学报: 自然科学版, 2014, 32(3): 252-259, 269. |
| Jing HY, Zhou Y, Guo XY, et al. Preliminary analysis of a Na+/H+ antiporter BgSOS1 at plasma membrane of Bruguiera gymnorrhiza[J]. Nat Sci J Hainan Univ, 2014, 32(3): 252-259, 269. | |
| [45] |
Gao JJ, Sun J, Cao PP, et al. Variation in tissue Na+ content and the activity of SOS1 genes among two species and two related Genera of Chrysanthemum[J]. BMC Plant Biol, 2016, 16: 98.
doi: 10.1186/s12870-016-0781-9 URL |
| [46] |
Venturini L, Ferrarini A, Zenoni S, et al. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity[J]. BMC Genomics, 2013, 14: 41.
doi: 10.1186/1471-2164-14-41 pmid: 23331995 |
| [47] |
Wang S, Li Z, Rui R, et al. Cloning and characterization of a plasma membrane Na+/H+ antiporter gene from Cucumis sativus[J]. Russ J Plant Physiol, 2013, 60(3): 330-336.
doi: 10.1134/S102144371303014X URL |
| [48] |
Hu J, Hu XK, Zhang HW, et al. Moderate NaCl alleviates osmotic stress in Lycium ruthenicum[J]. Plant Growth Regul, 2022, 96(1): 25-35.
doi: 10.1007/s10725-021-00754-0 |
| [49] |
Meng KB, Wu YX. Footprints of divergent evolution in two Na+/H+ type antiporter gene families(NHX and SOS1)in the genus Populus[J]. Tree Physiol, 2018, 38(6): 813-824.
doi: 10.1093/treephys/tpx173 URL |
| [50] | 李平, 王晓宇, 徐惠, 等. 蓖麻质膜型Na+/H+逆向转运蛋白基因(RcSOS1)克隆及表达载体构建[J]. 分子植物育种, 2018, 16(10): 3182-3189. |
| Li P, Wang XY, Xu H, et al. Cloning and expression vectors establishment of plasma membrane Na+/H+ antiporter gene(RcSOS1)in castor(Ricinus communis L.)[J]. Mol Plant Breed, 2018, 16(10): 3182-3189. | |
| [51] |
Li Q, Tang Z, Hu YB, et al. Functional analyses of a putative plasma membrane Na+/H+ antiporter gene isolated from salt tolerant Helianthus tuberosus[J]. Mol Biol Rep, 2014, 41(8): 5097-5108.
doi: 10.1007/s11033-014-3375-3 URL |
| [52] |
Guo JR, Dong XX, Han GL, et al. Salt-enhanced reproductive development of Suaeda salsa L. coincided with ion transporter gene upregulation in flowers and increased pollen K+ content[J]. Front Plant Sci, 2019, 10: 333.
doi: 10.3389/fpls.2019.00333 URL |
| [53] |
Wang WY, Liu YQ, Duan HR, et al. SsHKT1;1 is coordinated with SsSOS1 and SsNHX1 to regulate Na+ homeostasis in Suaeda salsa under saline conditions[J]. Plant Soil, 2020, 449(1/2): 117-131.
doi: 10.1007/s11104-020-04463-x |
| [54] | Wang HY, Tang XL, Shao CY, et al. Molecular cloning and bioinformatics analysis of a new plasma membrane Na+/H+ antiporter gene from the halophyte Kosteletzkya virginica[J]. Sci World J, 2014, 2014: 141675. |
| [55] |
Jarvis DE, Ryu CH, Beilstein MA, et al. Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula[J]. Mol Biol Evol, 2014, 31(8): 2094-2107.
doi: 10.1093/molbev/msu152 pmid: 24803640 |
| [56] | 马苏勇, 祝建波, 王爱英, 等. 大叶补血草质膜Na+/H+逆向转运蛋白基因SOS1的克隆及对番茄的遗传转化[J]. 生物技术通报, 2010(9): 111-115. |
| Ma SY, Zhu JB, Wang AY, et al. Cloning and genetic transformation of tomato with SOS1 gene from Limonium gmelinii[J]. Biotechnol Bull, 2010(9): 111-115. | |
| [57] |
Abd El-Moneim D, ELsarag EIS, Aloufi S, et al. Quinoa(Chenopodium quinoa Willd.): genetic diversity according to ISSR and SCoT markers, relative gene expression, and morpho-physiological variation under salinity stress[J]. Plants, 2021, 10(12): 2802.
doi: 10.3390/plants10122802 URL |
| [58] |
Ma Q, Li YX, Yuan HJ, et al. ZxSOS1 is essential for long-distance transport and spatial distribution of Na+ and K+ in the xerophyte Zygophyllum xanthoxylum[J]. Plant Soil, 2014, 374(1): 661-676.
doi: 10.1007/s11104-013-1891-x URL |
| [59] |
Wang Z, Hong YC, Li YM, et al. Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication[J]. Plant Biotechnol J, 2021, 19(1): 20-22.
doi: 10.1111/pbi.v19.1 URL |
| [60] |
Lu QH, Wang YQ, Xu JP, et al. Effect of ABA on physiological characteristics and expression of salt tolerance-related genes in Tartary buckwheat[J]. Acta Physiol Plant, 2021, 43(5): 1-11.
doi: 10.1007/s11738-020-03172-3 |
| [61] |
Yadav NS, Shukla PS, Jha A, et al. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco[J]. BMC Plant Biol, 2012, 12: 188.
doi: 10.1186/1471-2229-12-188 |
| [62] |
Yang Y, Xu LF, Li WX, et al. A Na+/H+ antiporter-encoding salt overly sensitive 1 gene, LpSOS1, involved in positively regulating the salt tolerance in Lilium pumilum[J]. Gene, 2023, 874: 147485.
doi: 10.1016/j.gene.2023.147485 URL |
| [63] | Kaewjiw N, Laksana C, Chanprame S. Cloning and identification of salt overly sensitive(SOS1)gene of sugarcane[J]. Int J Agric Biol, 2018, 20(7): 1569-1574. |
| [64] |
Han QQ, Wang YP, Li J, et al. The mechanistic basis of sodium exclusion in Puccinellia tenuiflora under conditions of salinity and potassium deprivation[J]. Plant J, 2022, 112(2): 322-338.
doi: 10.1111/tpj.v112.2 URL |
| [65] |
Núñez-Ramírez R, Sánchez-Barrena MJ, Villalta I, et al. Structural insights on the plant salt-overly-sensitive 1(SOS1)Na+/H+ antiporter[J]. J Mol Biol, 2012, 424(5): 283-294.
doi: 10.1016/j.jmb.2012.09.015 pmid: 23022605 |
| [66] |
Quintero FJ, Martinez-Atienza J, Villalta I, et al. Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1(SOS1)by phosphorylation of an auto-inhibitory C-terminal domain[J]. Proc Natl Acad Sci USA, 2011, 108(6): 2611-2616.
doi: 10.1073/pnas.1018921108 URL |
| [67] |
Goswami P, Paulino C, Hizlan D, et al. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1[J]. EMBO J, 2011, 30(2): 439-449.
doi: 10.1038/emboj.2010.321 URL |
| [68] |
Zheng M, Li JP, Zeng CW, et al. Subgenome-biased expression and functional diversification of a Na+/H+ antiporter homoeologs in salt tolerance of polyploid wheat[J]. Front Plant Sci, 2022, 13: 1072009.
doi: 10.3389/fpls.2022.1072009 URL |
| [69] |
Ma X, Li QH, Yu YN, et al. The CBL-CIPK pathway in plant response to stress signals[J]. Int J Mol Sci, 2020, 21(16): 5668.
doi: 10.3390/ijms21165668 URL |
| [70] |
Tang RJ, Wang C, Li KL, et al. The CBL-CIPK calcium signaling network: unified paradigm from 20 years of discoveries[J]. Trends Plant Sci, 2020, 25(6): 604-617.
doi: 10.1016/j.tplants.2020.01.009 URL |
| [71] |
Wu GQ, Wang JL, Li SJ. Genome-wide identification of Na+/H+ antiporter(NHX)genes in sugar beet(Beta vulgaris L.) and their regulated expression under salt stress[J]. Genes, 2019, 10(5): 401.
doi: 10.3390/genes10050401 URL |
| [72] |
Hao SH, Wang YR, Yan YX, et al. A review on plant responses to salt stress and their mechanisms of salt resistance[J]. Horticulturae, 2021, 7(6): 132.
doi: 10.3390/horticulturae7060132 URL |
| [73] |
Park HJ, Qiang Z, Kim WY, et al. Diurnal and circadian regulation of salt tolerance in Arabidopsis[J]. J Plant Biol, 2016, 59(6): 569-578.
doi: 10.1007/s12374-016-0317-8 URL |
| [74] |
Chen MX, She ZY, Aslam M, et al. Genomic insights of the WRKY genes in kenaf(Hibiscus cannabinus L.) reveal that HcWRKY44 improves the plant's tolerance to the salinity stress[J]. Front Plant Sci, 2022, 13: 984233.
doi: 10.3389/fpls.2022.984233 URL |
| [75] |
Wu X, Xu JN, Meng XN, et al. Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis[J]. Plant Physiol, 2022, 189(3): 1833-1847.
doi: 10.1093/plphys/kiac174 URL |
| [76] |
Zhang HF, Guo JB, Chen XQ, et al. Pepper bHLH transcription factor CabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis[J]. Hortic Res, 2022, 9: uhac203.
doi: 10.1093/hr/uhac203 URL |
| [77] |
Fu HQ, Yu X, Jiang YY, et al. SALT OVERLY SENSITIVE 1 is inhibited by clade D protein phosphatase 2C D6 and D7 in Arabidopsis thaliana[J]. Plant Cell, 2023, 35(1): 279-297.
doi: 10.1093/plcell/koac283 URL |
| [78] |
Duscha K, Martins Rodrigues C, Müller M, et al. 14-3-3 proteins and other candidates form protein-protein interactions with the cytosolic C-terminal end of SOS1 affecting its transport activity[J]. Int J Mol Sci, 2020, 21(9): 3334.
doi: 10.3390/ijms21093334 URL |
| [79] |
Kim WY, Ali Z, Park HJ, et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis[J]. Nat Commun, 2013, 4: 1352.
doi: 10.1038/ncomms2357 |
| [80] |
Li JF, Zhou HP, Zhang Y, et al. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana[J]. Dev Cell, 2020, 55(3): 367-380.e6.
doi: 10.1016/j.devcel.2020.08.005 URL |
| [81] |
Amirbakhtiar N, Ismaili A, Ghaffari MR, et al. Transcriptome analysis of bread wheat leaves in response to salt stress[J]. PLoS One, 2021, 16(7): e0254189.
doi: 10.1371/journal.pone.0254189 URL |
| [82] |
Zhou M, Wang W. SOS1 safeguards plant circadian rhythm against daily salt fluctuations[J]. Proc Natl Acad Sci USA, 2022, 119(36): e2212950119.
doi: 10.1073/pnas.2212950119 URL |
| [83] |
Pei SY, Liu YT, Li WK, et al. OSCA1 is an osmotic specific sensor: a method to distinguish Ca2+-mediated osmotic and ionic perception[J]. New Phytol, 2022, 235(4): 1665-1678.
doi: 10.1111/nph.v235.4 URL |
| [84] |
Yuan F, Yang HM, Xue Y, et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis[J]. Nature, 2014, 514(7522): 367-371.
doi: 10.1038/nature13593 |
| [85] |
Jiang ZH, Zhou XP, Tao M, et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx[J]. Nature, 2019, 572(7769): 341-346.
doi: 10.1038/s41586-019-1449-z |
| [86] |
Yin XC, Xia YQ, Xie Q, et al. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance[J]. J Exp Bot, 2020, 71(6): 1801-1814.
doi: 10.1093/jxb/erz549 URL |
| [87] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324.
doi: 10.1016/j.cell.2016.08.029 URL |
| [88] |
Guo Y, Liu Y, Zhang Y, et al. Effects of exogenous calcium on adaptive growth, photosynthesis, ion homeostasis and phenolics of Gleditsia sinensis Lam. plants under salt stress[J]. Agriculture, 2021, 11(10): 978.
doi: 10.3390/agriculture11100978 URL |
| [89] |
Fraile-Escanciano A, Kamisugi Y, Cuming AC, et al. The SOS1 transporter of Physcomitrella patens mediates sodium efflux in planta[J]. New Phytol, 2010, 188(3): 750-761.
doi: 10.1111/j.1469-8137.2010.03405.x pmid: 20696009 |
| [90] |
Gupta A, Shaw BP, Sahu BB. Post-translational regulation of the membrane transporters contributing to salt tolerance in plants[J]. Funct Plant Biol, 2021, 48(12): 1199-1212.
doi: 10.1071/FP21153 pmid: 34665998 |
| [91] |
Quintero FJ, Ohta M, Shi HZ, et al. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis[J]. Proc Natl Acad Sci USA, 2002, 99(13): 9061-9066.
doi: 10.1073/pnas.132092099 URL |
| [92] |
Zhou Y, Lai ZS, Yin XC, et al. Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco[J]. Plant Sci, 2016, 253: 176-186.
doi: 10.1016/j.plantsci.2016.09.016 URL |
| [93] |
Xie Q, Yang Y, Wang Y, et al. The calcium sensor CBL10 negatively regulates plasma membrane H+-ATPase activity and alkaline stress response in Arabidopsis[J]. Environ Exp Bot, 2022, 194: 104752.
doi: 10.1016/j.envexpbot.2021.104752 URL |
| [94] |
Park HJ, Kim WY, Yun DJ. A new insight of salt stress signaling in plant[J]. Mol Cells, 2016, 39(6): 447-459.
doi: 10.14348/molcells.2016.0083 pmid: 27239814 |
| [95] |
Du WM, Lin HX, Chen S, et al. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis[J]. Plant Physiol, 2011, 156(4): 2235-2243.
doi: 10.1104/pp.111.173377 URL |
| [96] |
谢玲玲, 王金龙, 伍国强. 植物CBL-CIPK信号系统响应非生物胁迫的调控机制[J]. 植物学报, 2021, 56(5): 614-626.
doi: 10.11983/CBB21024 |
| Xie LL, Wang JL, Wu GQ. Regulatory mechanisms of the plant CBL-CIPK signaling system in response to abiotic stress[J]. Chin Bull Bot, 2021, 56(5): 614-626. | |
| [97] |
Wang Q, Guan C, Wang P, et al. The effect of AtHKT1;1 or AtSOS1 mutation on the expressions of Na+ or K+ transporter genes and ion homeostasis in Arabidopsis thaliana under salt stress[J]. Int J Mol Sci, 2019, 20(5): 1085.
doi: 10.3390/ijms20051085 URL |
| [98] |
Venkataraman G, Shabala S, Véry AA, et al. To exclude or to accumulate? Revealing the role of the sodium HKT1;5 transporter in plant adaptive responses to varying soil salinity[J]. Plant Physiol Biochem, 2021, 169: 333-342.
doi: 10.1016/j.plaphy.2021.11.030 URL |
| [99] |
Riedelsberger J, Miller JK, Valdebenito-Maturana B, et al. Plant HKT channels: an updated view on structure, function and gene regulation[J]. Int J Mol Sci, 2021, 22(4): 1892.
doi: 10.3390/ijms22041892 URL |
| [100] | Gu WT, Zhou LB, Liu RY, et al. Synergistic responses of NHX, AKT1, and SOS1 in the control of Na+ homeostasis in sweet sorghum mutants induced by 12C6+-ion irradiation[J]. Nucl Sci Tech, 2018, 29(1): 10. |
| [101] |
Sun QA, Yamada T, Han YL, et al. Differential responses of NHX1 and SOS1 gene expressions to salinity in two Miscanthus sinensis anderss. accessions with different salt tolerance[J]. Phyton, 2021, 90(3): 827-836.
doi: 10.32604/phyton.2021.013805 URL |
| [102] |
Olías R, Eljakaoui Z, Li J, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs[J]. Plant Cell Environ, 2009, 32(7): 904-916.
doi: 10.1111/pce.2009.32.issue-7 URL |
| [103] |
Ishikawa T, Shabala L, Zhou MX, et al. Comparative analysis of root Na+ relation under salinity between Oryza sativa and Oryza coarctata[J]. Plants, 2022, 11(5): 656.
doi: 10.3390/plants11050656 URL |
| [104] |
Fan YF, Yin XC, Xie Q, et al. Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance[J]. BMC Plant Biol, 2019, 19(1): 74.
doi: 10.1186/s12870-019-1680-7 |
| [105] | Shabala S, Munns R. Salinity stress: physiological constraints and adaptive mechanisms[M]// Plant Stress Physiology. UK: CABI, 2017: 24-63. |
| [106] |
Katiyar-Agarwal S, Zhu JH, Kim K, et al. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis[J]. Proc Natl Acad Sci USA, 2006, 103(49): 18816-18821.
pmid: 17023541 |
| [107] |
Feki K, Tounsi S, Masmoudi K, et al. The durum wheat plasma membrane Na+/H+ antiporter SOS1 is involved in oxidative stress response[J]. Protoplasma, 2017, 254(4): 1725-1734.
doi: 10.1007/s00709-016-1066-8 URL |
| [108] |
Zhou Y, Yin XC, Wan SM, et al. The Sesuvium portulacastrum plasma membrane Na+/H+ antiporter SpSOS1 complemented the salt sensitivity of transgenic Arabidopsis sos1 mutant plants[J]. Plant Mol Biol Rep, 2018, 36(4): 553-563.
doi: 10.1007/s11105-018-1099-6 |
| [109] |
Xu XD, Yuan L, Xie QG. The circadian clock ticks in plant stress responses[J]. Stress Biol, 2022, 2(1): 15.
doi: 10.1007/s44154-022-00040-7 pmid: 37676516 |
| [110] |
Yang YQ, Wu YJ, Ma L, et al. The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis[J]. Plant Cell, 2019, 31(6): 1367-1384.
doi: 10.1105/tpc.18.00568 URL |
| [111] |
Oh DH, Lee SY, Bressan RA, et al. Intracellular consequences of SOS1 deficiency during salt stress[J]. J Exp Bot, 2010, 61(4): 1205-1213.
doi: 10.1093/jxb/erp391 URL |
| [112] |
Zhou JY, Hao DL, Yang GZ. Regulation of cytosolic pH: the contributions of plant plasma membrane H+-ATPases and multiple transporters[J]. Int J Mol Sci, 2021, 22(23): 12998.
doi: 10.3390/ijms222312998 URL |
| [113] | Guo KM, Babourina O, Rengel Z. Na+/H+ antiporter activity of the SOS1 gene: lifetime imaging analysis and electrophysiological studies on Arabidopsis seedlings[J]. Physiol Plant, 2009, 137(2): 155-165. |
| [114] |
Cha JY, Kim J, Jeong SY, et al. The Na+/H+ antiporter salt overly sensitive 1 regulates salt compensation of circadian rhythms by stabilizing gigantea in arabidopsis[J]. Proc Natl Acad Sci USA, 2022, 119(33): e2207275119.
doi: 10.1073/pnas.2207275119 URL |
| [115] |
Ma YC, Wang L, Wang JY, et al. Isolation and expression analysis of salt overly sensitive gene family in grapevine(Vitis vinifera)in response to salt and PEG stress[J]. PLoS One, 2019, 14(3): e0212666.
doi: 10.1371/journal.pone.0212666 URL |
| [116] |
Shahzad B, Yun P, Rasouli F, et al. Root K+ homeostasis and signalling as a determinant of salinity stress tolerance in cultivated and wild rice species[J]. Environ Exp Bot, 2022, 201: 104944.
doi: 10.1016/j.envexpbot.2022.104944 URL |
| [117] |
Chai HX, Guo JF, Zhong YL, et al. The plasma-membrane polyamine transporter PUT3 is regulated by the Na+/H+ antiporter SOS1 and protein kinase SOS2[J]. New Phytol, 2020, 226(3): 785-797.
doi: 10.1111/nph.v226.3 URL |
| [118] |
Farooq M, Park JR, Jang YH, et al. Rice cultivars under salt stress show differential expression of genes related to the regulation of Na+/K+ balance[J]. Front Plant Sci, 2021, 12: 680131.
doi: 10.3389/fpls.2021.680131 URL |
| [119] |
Yue YS, Zhang MC, Zhang JC, et al. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio[J]. J Plant Physiol, 2012, 169(3): 255-261.
doi: 10.1016/j.jplph.2011.10.007 URL |
| [120] |
Akrimi R, Hajlaoui H, Batelli G, et al. Electromagnetic water enhanced metabolism and agro-physiological responses of potato(Solanum tuberosum L)under saline conditions[J]. J Agron Crop Sci, 2021, 207(1): 44-58.
doi: 10.1111/jac.v207.1 URL |
| [121] |
Awaji SM, Hanjagi PS, Pushpa BN, et al. Overexpression of plasma membrane Na+/H+ antiporter OsSOS1 gene improves salt tolerance in transgenic rice plants[J]. Oryza, 2020, 57(4): 277-287.
doi: 10.35709/ory.2019.56.2 URL |
| [122] |
Rao YR, Ansari MW, Sahoo RK, et al. Salicylic acid modulates ACS, NHX1, sos1 and HKT1;2 expression to regulate ethylene overproduction and Na+ ions toxicity that leads to improved physiological status and enhanced salinity stress tolerance in tomato plants cv. Pusa Ruby[J]. Plant Signal Behav, 2021, 16(11): 1950888.
doi: 10.1080/15592324.2021.1950888 URL |
| [123] |
Chang PJ, Wu Z, Song NN, et al. Identification MdeSOS1 in Magnolia denudata and its function in response to salt stress[J]. J Plant Interact, 2020, 15(1): 417-426.
doi: 10.1080/17429145.2020.1843721 URL |
| [124] |
Hichri I, Muhovski Y, Clippe A, et al. SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and Arabidopsis[J]. Plant Cell Environ, 2016, 39(1): 62-79.
doi: 10.1111/pce.v39.1 URL |
| [1] | 康凌云, 韩露露, 韩德平, 陈建胜, 甘瀚凌, 邢凯, 马友记, 崔凯. 褪黑素缓解空肠黏膜上皮细胞氧化损伤的效果研究[J]. 生物技术通报, 2023, 39(9): 291-299. |
| [2] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [3] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [4] | 陈奕博, 杨万明, 岳爱琴, 王利祥, 杜维俊, 王敏. 基于SLAF标记的大豆遗传图谱构建及苗期耐盐性QTL定位[J]. 生物技术通报, 2023, 39(2): 70-79. |
| [5] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [6] | 高晓蓉, 丁尧, 吕军. 芘降解菌Pseudomonas sp. PR3的植物促生特性及其对芘胁迫下水稻生长的影响[J]. 生物技术通报, 2022, 38(9): 226-236. |
| [7] | 陈宏艳, 李小二, 李忠光. 糖信号及其在植物响应逆境胁迫中的作用[J]. 生物技术通报, 2022, 38(7): 80-89. |
| [8] | 石广成, 杨万明, 杜维俊, 王敏. 大豆耐盐种质的筛选及其耐盐生理特性分析[J]. 生物技术通报, 2022, 38(4): 174-183. |
| [9] | 张小妮, 翁伊纯, 范奕浩, 王晓娟, 赵佳宇, 张云龙. Mito-OS-Timer:一种靶向监测线粒体氧化应激的荧光秒表[J]. 生物技术通报, 2022, 38(10): 97-105. |
| [10] | 彭文超, 刘建新, 王迪铭. 哺乳动物低氧应激代谢成因的研究进展[J]. 生物技术通报, 2021, 37(1): 262-271. |
| [11] | 张晓佳, 卢亚军, 张文晋, 张瑜, 崔高畅, 郎多勇, 张新慧. 抗旱耐盐菌剂的制备及其对甘草种子萌发的影响[J]. 生物技术通报, 2020, 36(9): 180-193. |
| [12] | 鲁琳, 赵希胜, 刘维东, 王艳婷, 吴玲莉, 鲁黎明. 花烟草NáNHX1基因的克隆及其在非生物胁迫下的表达模式[J]. 生物技术通报, 2020, 36(4): 70-77. |
| [13] | 石晶静, 郭依萍, 于颖, 周美琪, 王超. 转BpLTP4烟草耐盐性分析[J]. 生物技术通报, 2020, 36(12): 34-41. |
| [14] | 刘融, 崔凯, 白福恒, 刁其玉. 蛋氨酸调控畜禽氧化应激的研究进展[J]. 生物技术通报, 2020, 36(10): 207-214. |
| [15] | 张昭杨, 庞军玲, 韩梅, 冷鹏飞, 赵军. 转基因ABP9玉米株系的耐盐性分析[J]. 生物技术通报, 2019, 35(5): 48-57. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||