








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 285-298.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1219
马想蓉( ), 马欣, 陈胤勋, 龙启福, 王嵘, 邢江娃(
), 马欣, 陈胤勋, 龙启福, 王嵘, 邢江娃( )
)
收稿日期:2023-12-29
出版日期:2024-07-26
发布日期:2024-05-24
通讯作者:
邢江娃,女,博士,副教授,研究方向:特殊环境微生物学;E-mail: xingjiangwa66@163.com作者简介:马想蓉,女,硕士研究生,研究方向:特殊环境微生物学;E-mail: 3480177451@qq.com
基金资助:
MA Xiang-rong( ), MA Xin, CHEN Yin-xun, LONG Qi-fu, WANG Rong, XING Jiang-wa(
), MA Xin, CHEN Yin-xun, LONG Qi-fu, WANG Rong, XING Jiang-wa( )
)
Received:2023-12-29
Published:2024-07-26
Online:2024-05-24
摘要:
【目的】 探究不同富集时间、培养盐度、稀释梯度、培养基等对氯化物型昆特依高盐盐湖可培养嗜盐细菌多样性的影响,确定嗜盐细菌的最佳分离培养条件,挖掘更多的环境嗜盐细菌资源。【方法】 从昆特依盐湖中采集水泥混合样本,选取7种培养基和10%、18% NaCl两个盐度,采用富集培养法、平板稀释涂布法和平板分区划线法分离嗜盐菌;通过16S rRNA基因测序与BLAST序列比对确定菌株系统分类学地位。同时采用免培养Illumina MiSeq高通量测序技术分析昆特依盐湖细菌群落结构多样性特征。【结果】 免培养昆特依盐湖样本中共鉴定到细菌39门64纲101目219科703属,其中假单胞菌门(Pseudomonadota)、厚壁菌门(Bacillota)、拟杆菌门(Bacteroidota)和放线菌门(Actinomycetota)为主要的优势菌门。根据菌落的大小、形态、颜色、光泽度、透明度、质地、隆起状态和边缘特征等,共分离出436株嗜盐菌,隶属于3门3纲6目9科28属77种,其中16株可能为潜在新种。3门为厚壁菌门(Bacillota)、假单胞菌门(Pseudomonadota)和放线菌门(Actinomycetota),且均为免培养测序结果中的优势菌门。在属水平上,芽孢杆菌属(Bacillus)、盐单胞菌属(Halomonas)、喜盐芽孢杆菌属(Halobacillus)、枝芽孢菌属(Virgibacillus)和卤水杆菌属(Salicola)为优势菌属。肿块芽孢杆菌属(Tuberibacillus)、越南蔷薇菌属(Rossellomorea)和谷氨酸杆菌属(Glutamicibacter)是首次从盐湖中分离得到。18% NaCl分离得到的菌株数量明显少于10% NaCl,且多样性较低,芽孢乳杆菌科(Sporolactobacillaceae)是该条件下分出的特有菌科。7种培养基中,1/2 RCA培养基分离效果最好。最佳富集培养时间为0、7和60 d,最佳样品稀释梯度为10-2和10-3。【结论】 从昆特依盐湖中共分离出嗜盐细菌3门3纲6目9科28属77种。应用多种培养基,设置不同盐度、富集培养时间和稀释梯度等可显著提升可培养嗜盐菌的多样性。
马想蓉, 马欣, 陈胤勋, 龙启福, 王嵘, 邢江娃. 柴达木盆地昆特依盐湖可培养嗜盐细菌多样性研究[J]. 生物技术通报, 2024, 40(7): 285-298.
MA Xiang-rong, MA Xin, CHEN Yin-xun, LONG Qi-fu, WANG Rong, XING Jiang-wa. Diversity of Culturable Halophilic Bacteria in the Chloride Type Kunteyi Salt Lake in the Qaidam Basin[J]. Biotechnology Bulletin, 2024, 40(7): 285-298.
| 样品 Sample | Sobs指数 Sobs index | Chao指数 Chao index | Ace指数 Ace index | Shannon指数 Shannon index | Simpson指数 Simpson index | Coverage指数 Coverage index |
|---|---|---|---|---|---|---|
| KS1 | 1 018 | 1 204.62 | 1 230.55 | 1.72 | 0.602 | 0.99 |
| KS2 | 968 | 1 384.02 | 1 125.13 | 4.74 | 0.033 | 0.99 |
| KS3 | 1 341 | 1 502.00 | 1 418.44 | 5.89 | 0.008 | 1.00 |
| KS4 | 1 178 | 1 448.01 | 1 321.52 | 3.84 | 0.127 | 0.99 |
表1 昆特依盐湖样本中的细菌群落α多样性指数
Table 1 Alpha diversity index of bacterial communities in samples from Kunteyi Salt Lake
| 样品 Sample | Sobs指数 Sobs index | Chao指数 Chao index | Ace指数 Ace index | Shannon指数 Shannon index | Simpson指数 Simpson index | Coverage指数 Coverage index |
|---|---|---|---|---|---|---|
| KS1 | 1 018 | 1 204.62 | 1 230.55 | 1.72 | 0.602 | 0.99 |
| KS2 | 968 | 1 384.02 | 1 125.13 | 4.74 | 0.033 | 0.99 |
| KS3 | 1 341 | 1 502.00 | 1 418.44 | 5.89 | 0.008 | 1.00 |
| KS4 | 1 178 | 1 448.01 | 1 321.52 | 3.84 | 0.127 | 0.99 |
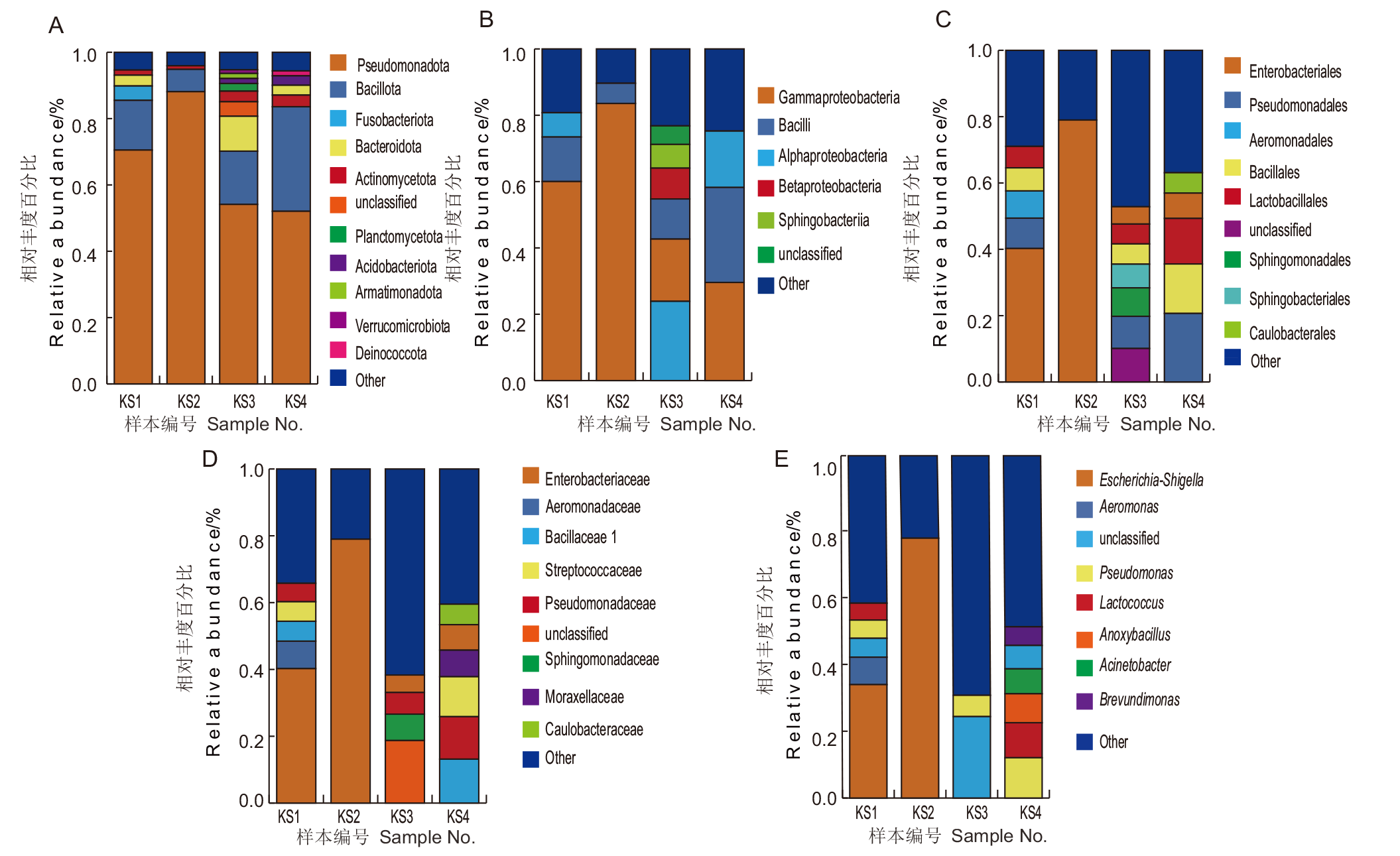
图1 昆特依盐湖免培养细菌多样性分析 A:门水平;B:纲水平;C:目水平;D:科水平;E:属水平
Fig. 1 Diversity analysis of culture-free bacteria in Kunteyi Salt Lake A: Phylum. B: Class. C: Order. D: Family. E: Genus
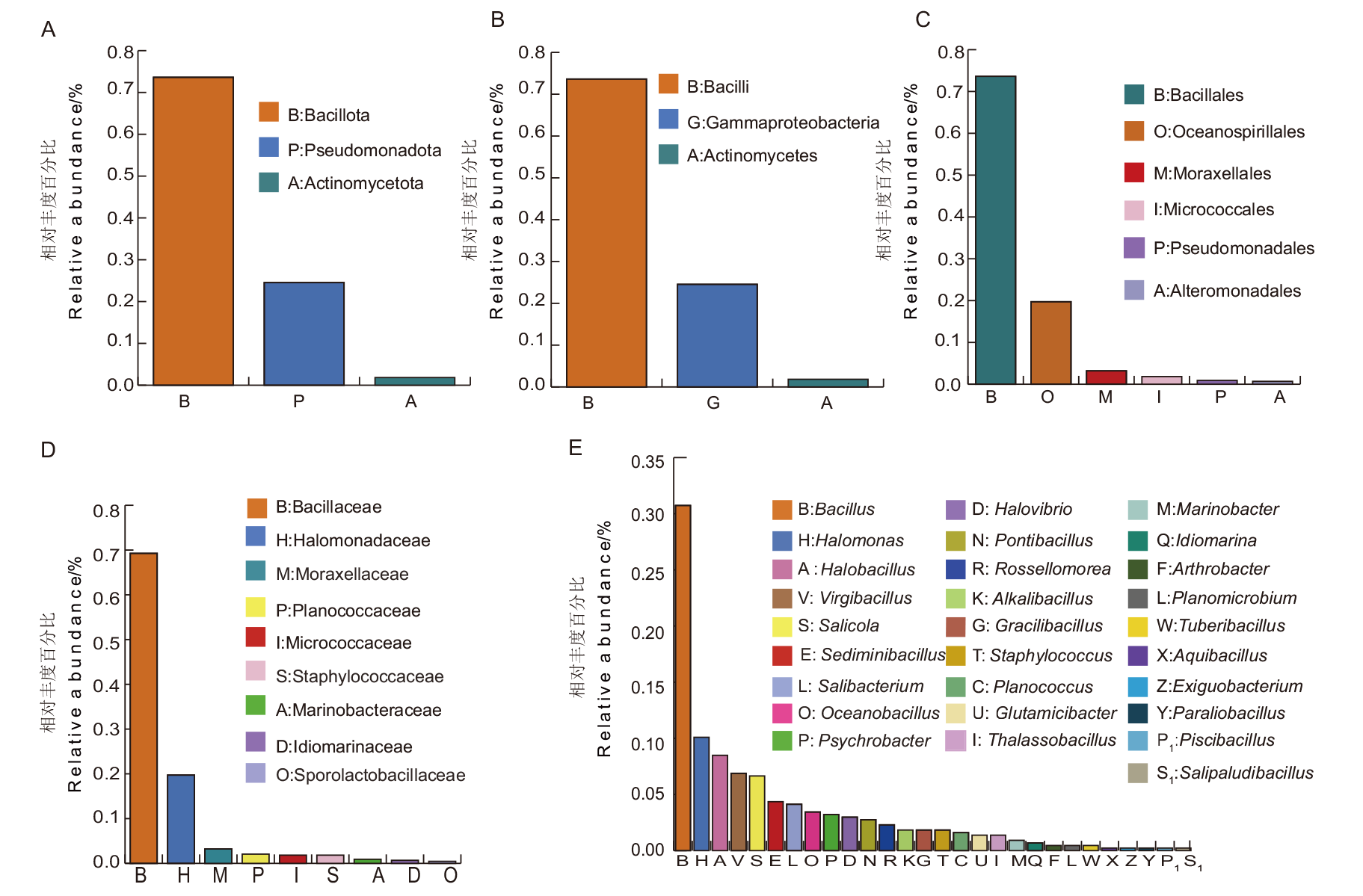
图2 昆特依盐湖可培养嗜盐细菌多样性分析 A:门水平;B:纲水平;C:目水平;D:科水平;E:属水平
Fig. 2 Diversity analysis of culturable halophilic bacteria in Kunteyi Salt Lake A: Phylum. B: Class. C: Order. D: Family. E: Genus
| 培养基 Medium | 菌株数量 Number of strains | 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species | 优势度指数Dominance index |
|---|---|---|---|---|---|---|---|---|
| 1/10 2216E | 39 | 2 | 2 | 3 | 3 | 13 | 17 | 0.22 |
| 2216E | 50 | 2 | 2 | 3 | 3 | 12 | 17 | 0.22 |
| 1/2 R2A | 76 | 2 | 2 | 2 | 3 | 15 | 34 | 0.44 |
| 1/2 RCA | 99 | 3 | 3 | 5 | 7 | 16 | 27 | 0.35 |
| 1/2 TSA | 51 | 2 | 2 | 3 | 5 | 11 | 22 | 0.29 |
| OSM | 77 | 2 | 2 | 4 | 6 | 12 | 16 | 0.21 |
| MGM | 44 | 2 | 2 | 2 | 2 | 12 | 17 | 0.22 |
表2 不同培养基可分离嗜盐菌多样性
Table 2 Biodiversity of halophilic bacteria isolated by different media
| 培养基 Medium | 菌株数量 Number of strains | 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species | 优势度指数Dominance index |
|---|---|---|---|---|---|---|---|---|
| 1/10 2216E | 39 | 2 | 2 | 3 | 3 | 13 | 17 | 0.22 |
| 2216E | 50 | 2 | 2 | 3 | 3 | 12 | 17 | 0.22 |
| 1/2 R2A | 76 | 2 | 2 | 2 | 3 | 15 | 34 | 0.44 |
| 1/2 RCA | 99 | 3 | 3 | 5 | 7 | 16 | 27 | 0.35 |
| 1/2 TSA | 51 | 2 | 2 | 3 | 5 | 11 | 22 | 0.29 |
| OSM | 77 | 2 | 2 | 4 | 6 | 12 | 16 | 0.21 |
| MGM | 44 | 2 | 2 | 2 | 2 | 12 | 17 | 0.22 |
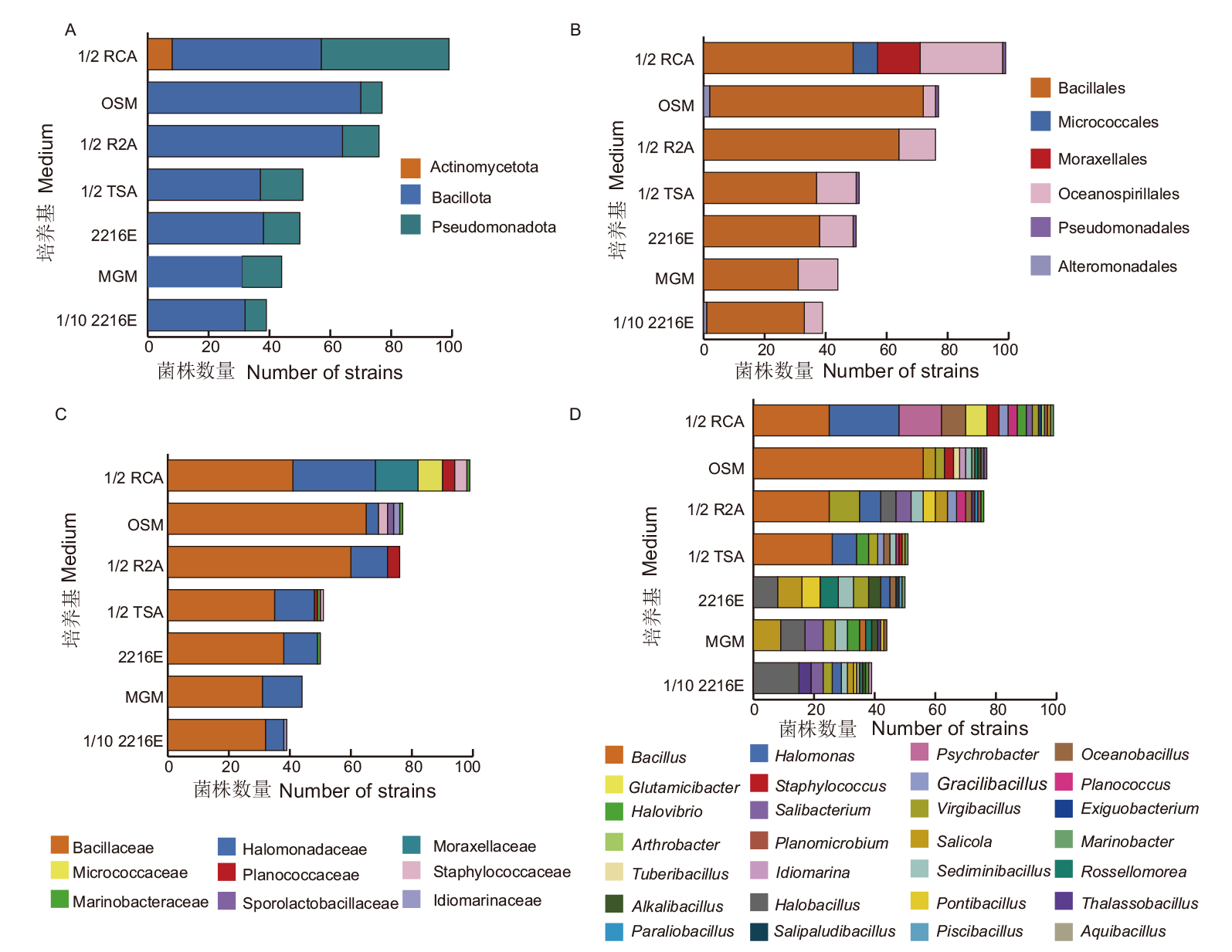
图4 不同培养基嗜盐菌在门(A)、目(B)、科(C)、属(D)水平上的差异性
Fig. 4 Differences of halophilic bacteria isolated by different media at the level of phylum(A), order(B), family(C), and genus(D)
| 富集时期 Enrichment period/d | 菌株数量 Number of strains | 比例 Scale/% |
|---|---|---|
| 0 | 77 | 17.66 |
| 7 | 56 | 12.84 |
| 14 | 53 | 12.16 |
| 21 | 68 | 15.60 |
| 30 | 78 | 17.89 |
| 45 | 56 | 12.84 |
| 60 | 48 | 11.01 |
表3 不同富集时期分离菌株数量
Table 3 Number of bacterial strains isolated at different enrichment periods
| 富集时期 Enrichment period/d | 菌株数量 Number of strains | 比例 Scale/% |
|---|---|---|
| 0 | 77 | 17.66 |
| 7 | 56 | 12.84 |
| 14 | 53 | 12.16 |
| 21 | 68 | 15.60 |
| 30 | 78 | 17.89 |
| 45 | 56 | 12.84 |
| 60 | 48 | 11.01 |
| 富集时期 Enrichment period/d | 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species |
|---|---|---|---|---|---|---|
| 0 | 2 | 2 | 3 | 5 | 16 | 29 |
| 7 | 3 | 3 | 4 | 6 | 16 | 25 |
| 14 | 2 | 2 | 2 | 3 | 14 | 20 |
| 21 | 2 | 2 | 3 | 5 | 13 | 21 |
| 30 | 2 | 2 | 2 | 4 | 16 | 28 |
| 45 | 2 | 2 | 3 | 4 | 14 | 22 |
| 60 | 2 | 2 | 4 | 4 | 13 | 24 |
表4 不同富集时期可培养嗜盐菌多样性统计
Table 4 Culturable halophilic bacterial diversity statistics at different enrichment periods
| 富集时期 Enrichment period/d | 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species |
|---|---|---|---|---|---|---|
| 0 | 2 | 2 | 3 | 5 | 16 | 29 |
| 7 | 3 | 3 | 4 | 6 | 16 | 25 |
| 14 | 2 | 2 | 2 | 3 | 14 | 20 |
| 21 | 2 | 2 | 3 | 5 | 13 | 21 |
| 30 | 2 | 2 | 2 | 4 | 16 | 28 |
| 45 | 2 | 2 | 3 | 4 | 14 | 22 |
| 60 | 2 | 2 | 4 | 4 | 13 | 24 |
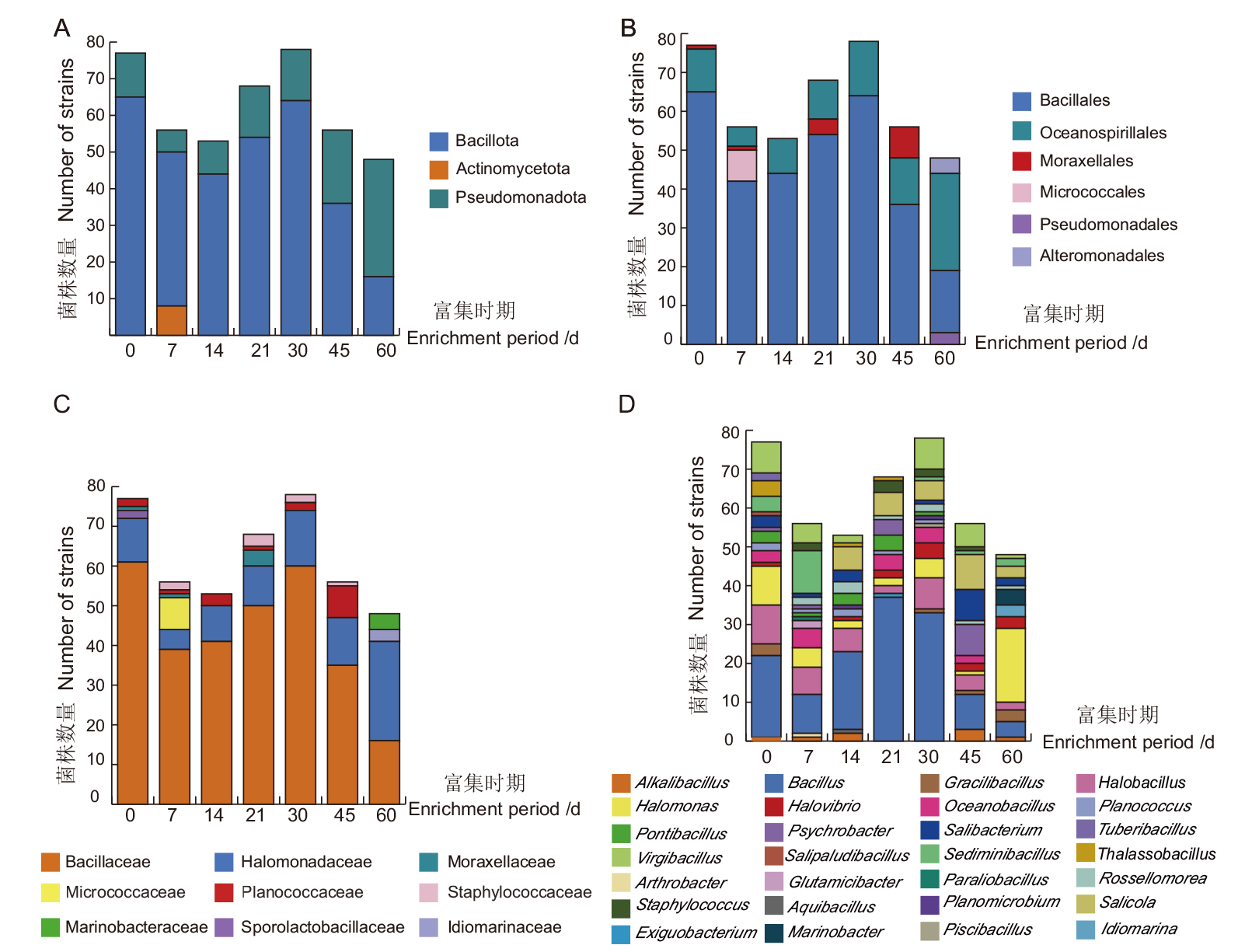
图5 不同富集时期嗜盐菌在门(A)、目(B)、科(C)、属(D)水平上的差异性
Fig. 5 Differences of halophilic bacteria at different enrichment periods at the level of phylum(A), order(B), family(C)and genus(D)
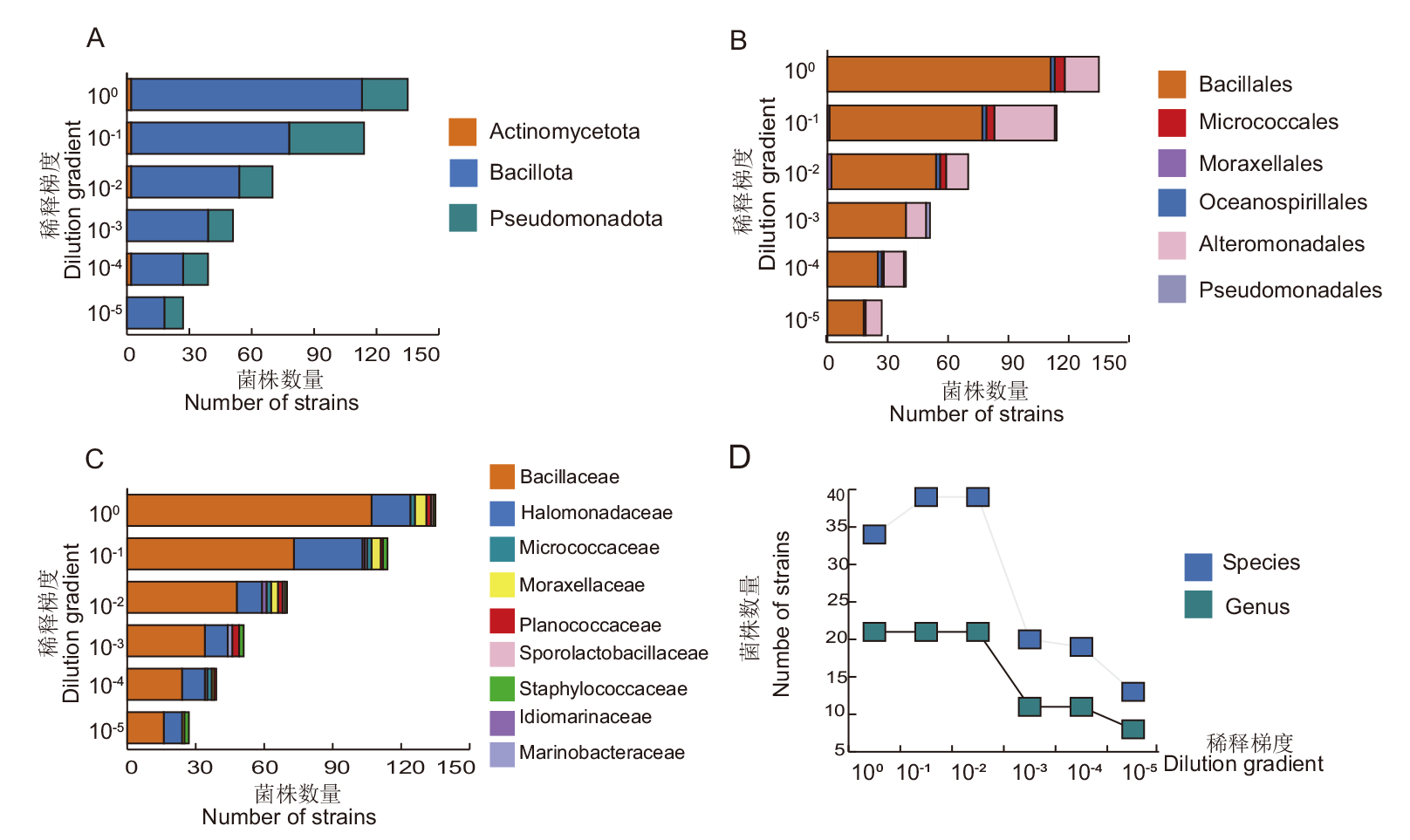
图7 不同稀释梯度嗜盐菌在门(A)、目(B)、科(C)、属和种(D)水平上的多样性
Fig. 7 Diversity of halophilic bacteria isolated by different dilution gradients at phylum(A),order(B), family(C), genus and species(D)levels
| 菌株编号 Strain No. | 近缘菌株 Phylogenetically related strain | 16S rRNA基因相似度 16S rRNA gene similarity/% |
|---|---|---|
| KTR1161 | Halomonas vilamensis SV325 | 98.56 |
| KTR1102 | Salibacterium salarium DSM 16461 | 98.55 |
| KTT1052 | Sediminibacillus halophilus T346 | 98.48 |
| KTF2041 | Halobacillus virgiliensis S3 | 98.48 |
| KTT4031 | Bacillus licheniformis CICC | 98.46 |
| KTT5052 | Bacillus licheniformis ALB | 98.45 |
| KTE2041 | Piscibacillus halophilus AT3RS18 | 98.27 |
| KTC1131 | Halovibrio denitrificans HGD 3 | 98.19 |
| KTO0101 | Tuberibacillus calidus 607 | 98.30 |
| KTE1061 | Marinobacter aquaticus M6-53 | 98.09 |
| KTT2052 | Halovibrio mesolongii HMY2 | 97.92 |
| KTF2021 | Aquibacillus koreensis BH30097 | 97.55 |
| KTO1051 | Bacillus haynesii INH14A | 97.45 |
| KTR3031 | Bacillus licheniformis F2 | 97.38 |
| KTR2043 | Virgibacillus byunsanensis ISL-24 | 97.35 |
| KTF3021 | Alkalibacillus aidingensis YIM 98829 | 96.94 |
表5 昆特依盐湖疑似新种
Table 5 Suspected new species in Kunteyi Salt Lake
| 菌株编号 Strain No. | 近缘菌株 Phylogenetically related strain | 16S rRNA基因相似度 16S rRNA gene similarity/% |
|---|---|---|
| KTR1161 | Halomonas vilamensis SV325 | 98.56 |
| KTR1102 | Salibacterium salarium DSM 16461 | 98.55 |
| KTT1052 | Sediminibacillus halophilus T346 | 98.48 |
| KTF2041 | Halobacillus virgiliensis S3 | 98.48 |
| KTT4031 | Bacillus licheniformis CICC | 98.46 |
| KTT5052 | Bacillus licheniformis ALB | 98.45 |
| KTE2041 | Piscibacillus halophilus AT3RS18 | 98.27 |
| KTC1131 | Halovibrio denitrificans HGD 3 | 98.19 |
| KTO0101 | Tuberibacillus calidus 607 | 98.30 |
| KTE1061 | Marinobacter aquaticus M6-53 | 98.09 |
| KTT2052 | Halovibrio mesolongii HMY2 | 97.92 |
| KTF2021 | Aquibacillus koreensis BH30097 | 97.55 |
| KTO1051 | Bacillus haynesii INH14A | 97.45 |
| KTR3031 | Bacillus licheniformis F2 | 97.38 |
| KTR2043 | Virgibacillus byunsanensis ISL-24 | 97.35 |
| KTF3021 | Alkalibacillus aidingensis YIM 98829 | 96.94 |
| [1] | 庄滢潭, 刘芮存, 陈雨露, 等. 极端微生物及其应用研究进展[J]. 中国科学: 生命科学, 2022, 52(2): 204-222. |
| Zhuang YT, Liu RC, Chen YL, et al. Extremophiles and their applications[J]. Sci Sin Vitae, 2022, 52(2): 204-222. | |
| [2] | Sánchez-Porro C. Special issue “halophilic microorganisms”[J]. Microorganisms, 2023, 11(3): 690. |
| [3] |
Pikuta EV, Hoover RB, Tang J. Microbial extremophiles at the limits of life[J]. Crit Rev Microbiol, 2007, 33(3): 183-209.
pmid: 17653987 |
| [4] | Mitra R, Xu T, Xiang H, et al. Current developments on polyhydroxyalkanoates synthesis by using halophiles as a promising cell factory[J]. Microb Cell Fact, 2020, 19(1): 86. |
| [5] | Dutta B, Bandopadhyay R. Biotechnological potentials of halophilic microorganisms and their impact on mankind[J]. Beni Suef Univ J Basic Appl Sci, 2022, 11(1): 75. |
| [6] | 刘妮娜, 崔甜琦, 高翔, 等. 嗜盐微生物在生物医药领域的应用研究进展[J]. 生物工程学报, 2022, 38(6): 2153-2168. |
| Liu NN, Cui TQ, Gao X, et al. Advances in the biomedical application research of halophilic microorganisms[J]. Chin J Biotechnol, 2022, 38(6): 2153-2168. | |
| [7] | 杨游胜, 姚智豪, 赵志星, 等. 富锂硫酸盐型盐湖卤水蒸发实验研究进展[J/OL]. 无机盐工业, 2023. DOI: 10.19964/j.issn.1006-4990.2023-0481. |
| Yang YS, Yao ZH, Zhao ZX, et al. Research progress of lithium-rich sulfate type salt lake brine evaporation experiment[J/OL]. Inorganic Chemicals Industry, 2023. DOI: 10.19964/j.issn.1006-4990.2023-0481. | |
| [8] | Li C, Chen XQ, Guo HF, et al. Production of potash and N-Mg compound fertilizer via mineral shoenite from kunteyi salt lake: phase diagrams of quaternary system(NH4)2SO4-MgSO4-K2SO4-H2O in the isothermal evaporation and crystallization process[J]. Acta Geol Sin Engl Ed, 2021, 95(3): 1016-1023. |
| [9] | 沈国平, 韩睿, 缪增强, 等. 青藏高原4类典型水化学特征湖泊的细菌多样性差异及影响因素[J]. 生物多样性, 2022, 30(4): 88-102. |
| Shen GP, Han R, Miao ZQ, et al. Bacterial diversity differences and influence factors of four types of hydrochemical characteristic lakes in the Qinghai-Tibet Plateau[J]. Biodivers Sci, 2022, 30(4): 88-102. | |
| [10] | Jacob JH, Hussein EI, Ali K Shakhatreh M, et al. Microbial community analysis of the hypersaline water of the Dead Sea using high-throughput amplicon sequencing[J]. MicrobiologyOpen, 2017, 6(5): e00500. |
| [11] |
Martínez JM, Escudero C, Rodríguez N, et al. Subsurface and surface halophile communities of the chaotropic Salar de Uyuni[J]. Environ Microbiol, 2021, 23(7): 3987-4001.
doi: 10.1111/1462-2920.15411 pmid: 33511754 |
| [12] | 沈硕. 青藏高原察尔汗盐湖地区可培养中度嗜盐菌的群落结构与多样性[J]. 微生物学报, 2017, 57(4): 490-499. |
| Shen S. Community structure and diversity of culturable moderate halophilic bacteria isolated from Qrhan Salt Lake on Qinghai-Tibet Plateau[J]. Acta Microbiol Sin, 2017, 57(4): 490-499. | |
| [13] | 马喆, 韩凤清, 易磊, 等. 柴达木盆地昆特依盐湖沉积特征及其盐类资源评价[J]. 盐湖研究, 2020, 28(1): 86-95. |
| Ma Z, Han FQ, Yi L, et al. Sedimentary characteristics and assessment of salt mineral resources in kunteyi salt lake, Qaidam Basin[J]. J Salt Lake Res, 2020, 28(1): 86-95. | |
| [14] | 周留艳. 基于代谢组学的海洋难培养微生物分离培养研究[D]. 济南: 山东大学, 2020. |
| Zhou LY. Study on the separation and cultivation of yet-to-be cultured marine microorganism based on metabolomics[D]. Jinan: Shandong University, 2020. | |
| [15] | 贺瑞含. 南北极环境样品微生物多样性分析、病原拮抗菌筛选及新物种的鉴定[D]. 济南: 山东大学, 2019. |
| He RH. Bacterial diversity analysis of Arctic and Antarctic environmental samples, screening of pathogen-antagonistic bacteria and identification of novel species[D]. Jinan: Shandong University, 2019. | |
| [16] | 李佳丽, 陈稳迪, 冯广达, 等. 广东南岭森林土壤中可培养细菌多样性[J]. 生物资源, 2020, 42(5): 593-601. |
| Li JL, Chen WD, Feng GD, et al. Diversity of culturable bacteria isolated from forest soils in the Nanling National Nature Reserve, Guangdong province[J]. Biotic Resour, 2020, 42(5): 593-601. | |
| [17] | 孙创, 王金燕, 张钰琳, 等. 利用改良培养基探究西太平洋海水可培养细菌多样性[J]. 微生物学报, 2021, 61(4): 845-861. |
| Sun C, Wang JY, Zhang YL, et al. Exploring the diversity of cultivated bacteria in the Western Pacific waters through improved culture media[J]. Acta Microbiol Sin, 2021, 61(4): 845-861. | |
| [18] |
张田田, 李永臻, 沈国平, 等. 高盐盐湖可分离嗜盐耐盐菌的种群多样性及四氢嘧啶产量评价[J]. 生物技术通报, 2022, 38(1): 168-178.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0201 |
| Zhang TT, Li YZ, Shen GP, et al. Population diversity of isolated halophilic and halotolerant bacteria from hypersaline salt lakes and evaluation of ectoine production[J]. Biotechnol Bull, 2022, 38(1): 168-178. | |
| [19] | 王旭红. 嗜盐古生菌Natrinema sp. J7-2的表层蛋白功能的初步研究[D]. 武汉: 武汉大学, 2018. |
| Wang XH. Preliminary study on the function of cell surface proteins in haloarchaeon Natrinema sp. J7-2[D]. Wuhan: Wuhan University, 2018. | |
| [20] | 程刚, 程艳, 胡海洋, 等. 伊犁河流域春季可培养细菌多样性及其胞外酶检测[J]. 微生物学杂志, 2023, 43(4): 36-48. |
| Cheng G, Cheng Y, Hu HY, et al. Diversity characteristics of cultivable bacteria and screening of enzyme-producing strains in Ili River Basin in spring[J]. J Microbiol, 2023, 43(4): 36-48. | |
| [21] | 中华人民共和国教育部. 离子色谱分析方法通则: JY/T 0575—2020[S]. 北京: 2020. |
| Ministry of Education of the People's Republic of China. General rules of analytical methods for ion chromatography: JY/T 0575—2020[S]. Beijing: 2020. | |
| [22] | Kim M, Oh HS, Park SC, et al. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes[J]. Int J Syst Evol Microbiol, 2014, 64(Pt 2): 346-351. |
| [23] | Pédron J, Guyon L, Lecomte A, et al. Comparison of environmental and culture-derived bacterial communities through 16S metabarcoding: a powerful tool to assess media selectivity and detect rare taxa[J]. Microorganisms, 2020, 8(8): 1129. |
| [24] | 田蕾, 李恩源, 关统伟, 等. 艾丁湖可培养嗜盐菌多样性及功能酶、抗菌活性筛选[J]. 微生物学通报, 2017, 44(11): 2575-2587. |
| Tian L, Li EY, Guan TW, et al. Diversity and functional enzymes, antimicrobial activity screening of culturable halophilic bacteria from Aiding Lake[J]. Microbiol China, 2017, 44(11): 2575-2587. | |
| [25] |
Borsodi AK, Kiss RI, Cech G, et al. Diversity and activity of cultivable aerobic planktonic bacteria of a saline Lake located in Sovata, Romania[J]. Folia Microbiol, 2010, 55(5): 461-466.
doi: 10.1007/s12223-010-0077-7 pmid: 20941581 |
| [26] |
Kajale S, Deshpande N, Shouche Y, et al. Cultivation of diverse microorganisms from hypersaline lake and impact of delay in sample processing on cell viability[J]. Curr Microbiol, 2020, 77(5): 716-721.
doi: 10.1007/s00284-019-01857-8 pmid: 31912221 |
| [27] | Meena D, Pandey J, Vasita R, et al. Genome sequence of Salipaludibacillus sp. strain CUR1, isolated from an alkaline-saline lake in Rajasthan, India[J]. Microbiol Resour Announc, 2022, 11(7): e0109221. |
| [28] | Singh H, Kaur M, Singh S, et al. Salibacterium nitratireducens sp. nov., a haloalkalitolerant bacterium isolated from a water sample from Sambhar salt lake, India[J]. Int J Syst Evol Microbiol, 2018, 68(11): 3506-3511. |
| [29] | Didari M, Bagheri M, Ali Amoozegar M, et al. Diversity of halophilic and halotolerant bacteria in the largest seasonal hypersaline lake(Aran-Bidgol-Iran)[J]. J Environ Health Sci Eng, 2020, 18(2): 961-971. |
| [30] | Amoozegar MA, Bagheri M, Didari M, et al. Aquibacillus halophilus gen. nov., sp. nov., a moderately halophilic bacterium from a hypersaline lake, and reclassification of Virgibacillus koreensis as Aquibacillus koreensis comb. nov. and Virgibacillus albus as Aquibacillus albus comb. nov[J]. Int J Syst Evol Microbiol, 2014, 64(Pt 11): 3616-3623. |
| [31] | 张莹, 石萍, 马炯. 微小杆菌Exiguobacterium spp.及其环境应用研究进展[J]. 应用与环境生物学报, 2013, 19(5):898-904. |
| Zhang Y, Shi P, Ma J. Exiguobacterium spp. and their applications in environmental remediation[J]. Chin J Appl Environ Biol, 2013, 19(05):898-904. | |
| [32] | 杨珊珊, 张晓波, 陈锋, 等. 新疆极端盐碱环境巴里坤湖沉积土壤中细菌的分离培养和鉴定[J]. 疾病预防控制通报, 2019, 34(5): 1-4, 16. |
| Yang SS, Zhang XB, Chen F, et al. Culture, isolation and identification of bacteria from sedimentary soil in environment of extreme saline alkali in Balikun Lake of Xinjiang[J]. Bull Dis Contr Prev China, 2019, 34(5): 1-4, 16. | |
| [33] | 董磊, 李姜维, 杨彩云, 等. 红树林土壤中脂肪酶产生菌的筛选及酶学性质[J]. 厦门大学学报: 自然科学版, 2010, 49(4): 570-573. |
| Dong L, Li JW, Yang CY, et al. Screening of lipase-producing bacteria from mangrove soil and preliminary studies on properties of enzyme[J]. J Xiamen Univ Nat Sci, 2010, 49(4): 570-573. | |
| [34] | 雷芬芬. 深海菌株Exiguobacterium sp. SWJS2发酵产蛋白酶、酶学性质及酶解特异性研究[D]. 广州: 华南理工大学, 2014. |
| Lei FF. Purification, characterization and hydrolysis specificity of protease from deep-sea strain Exiguobacterium sp. SWJS2[D]. Guangzhou: South China University of Technology, 2014. | |
| [35] | 夏晓敏, 汪建君, 陈立奇, 等. 厦门市区10月份大气气溶胶中细菌群落结构的初步研究[J]. 厦门大学学报: 自然科学版, 2010, 49(5): 682-687. |
| Xia XM, Wang JJ, Chen LQ, et al. Preliminary study of airborne bacteria community structure of aerosols of Xiamen in October[J]. J Xiamen Univ Nat Sci, 2010, 49(5): 682-687. | |
| [36] | Trilokesh C, Harish BS, Uppuluri KB. The antibiofilm potential of a heteropolysaccharide produced and characterized from the isolated marine bacterium Glutamicibacter nicotianae BPM30[J]. Prep Biochem Biotechnol, 2024, 54(2): 175-183. |
| [37] |
Tang L, Liu HX, Sun WN, et al. Halovibrio salipaludis sp. nov., isolated from saline-alkaline soil[J]. Curr Microbiol, 2021, 78(1): 429-434.
doi: 10.1007/s00284-020-02282-y pmid: 33219431 |
| [38] | John J, Siva V, Kumari R, et al. Unveiling cultivable and uncultivable halophilic bacteria inhabiting marakkanam saltpan, India and their potential for biotechnological applications[J]. Geomicrobiol J, 2020, 37(8): 691-701. |
| [39] | Ramezani M, Nikou MM, Pourmohyadini M, et al. Planomicrobium iranicum sp. nov., a novel slightly halophilic bacterium isolated from a hypersaline wetland[J]. Int J Syst Evol Microbiol, 2019, 69(5): 1433-1437. |
| [40] | Wang X, Wang Z, Zhao XQ, et al. Planococcus ruber sp. nov., isolated from a polluted farmland soil sample[J]. Int J Syst Evol Microbiol, 2017, 67(8): 2549-2554. |
| [41] | Akbari E, Beheshti-Maal K, Nayeri H. Production and optimization of alkaline lipase by a novel psychrotolerant and halotolerant strain Planomicrobium okeanokoites ABN-IAUF-2 isolated from Persian Gulf[J]. Int J Med Res Health Sci, 2016, 5: 139-148. |
| [42] | 努斯热提古丽·安外尔. 北疆胡杨茎秆液及胡杨碱的微生物多样性分析和植物促生菌的筛选[D]. 乌鲁木齐: 新疆大学, 2018. |
| Nusratgul A. Bacterial diversity of storage liquid and sap from populus euphratic in northern Xinjiang and screening of plant-promoting bacteria[D]. Urumqi: Xinjiang University, 2018. | |
| [43] | Yin QJ, Tang HZ, Zhu FC, et al. Complete genome of Rossellomorea sp. DA94, an agarolytic and orange-pigmented bacterium isolated from mangrove sediment of the South China Sea[J]. Mar Genomics, 2023, 71: 101059. |
| [44] | Dat TTH, Oanh PTT, Cuong LCV, et al. Pharmacological properties, volatile organic compounds, and genome sequences of bacterial endophytes from the mangrove plant Rhizophora apiculata blume[J]. Antibiotics, 2021, 10(12): 1491. |
| [45] | Navarro-Torre S, Carro L, Igual JM, et al. Rossellomorea arthrocnemi sp. nov., a novel plant growth-promoting bacterium used in heavy metal polluted soils as a phytoremediation tool[J]. Int J Syst Evol Microbiol, 2021, 71(10): 005015. |
| [46] | Khamwan S, Boonlue S, Mongkolthanaruk W. Production of fructan synthesis/hydrolysis of endophytic bacteria involved in inulin production in Jerusalem artichoke[J]. 3 Biotech, 2022, 12(11): 296. |
| [47] | 刘波, 陈倩倩, 阮传清, 等. 养猪微生物发酵床芽胞杆菌空间生态位特性[J]. 生态学报, 2019, 39(11): 4168-4189. |
| Liu B, Chen QQ, Ruan CQ, et al. Characteristics of the spatial ecological niche of Bacillus-like Genera in a microbial fermentation bed used for pig raising[J]. Acta Ecol Sin, 2019, 39(11): 4168-4189. | |
| [48] | Han WJ, Chen SN, Tan X, et al. Microbial community succession in response to sludge composting efficiency and heavy metal detoxification during municipal sludge composting[J]. Front Microbiol, 2022, 13: 1015949. |
| [49] | Mu DS, Liang QY, Wang XM, et al. Metatranscriptomic and comparative genomic insights into resuscitation mechanisms during enrichment culturing[J]. Microbiome, 2018, 6(1): 230. |
| [50] | 张科, 李臻, 郑瑶, 等. 河南叶县岩盐可培养中度嗜盐菌的多样性[J]. 微生物学通报, 2020, 47(12): 3987-3997. |
| Zhang K, Li Z, Zheng Y, et al. Biodiversity of culturable moderate halophilic bacteria of rock salt in Yexian County, Henan Province[J]. Microbiol China, 2020, 47(12): 3987-3997. | |
| [51] | 刘萍, 钱嘉兴, 谢连欢, 等. 具有群体感应抑制活性的红树林底泥海洋放线菌的分离筛选与鉴定[J]. 工业微生物, 2023, 53(4): 164-169. |
| Liu P, Qian JX, Xie LH, et al. Isolation, screening and identification of marine actinomycetes from mangrove sediments with quorum sensing inhibitory activity[J]. Ind Microbiol, 2023, 53(4): 164-169. |
| [1] | 於莉军, 王桥美, 彭文书, 严亮, 杨瑞娟. 景迈山古茶园与现代有机茶园根际土壤微生物群落研究[J]. 生物技术通报, 2024, 40(5): 237-247. |
| [2] | 雷美玲, 饶文华, 胡进锋, 岳琪, 吴祖建, 范国成. 黄龙病发病芦柑根际土壤细菌群落组成与多样性特征[J]. 生物技术通报, 2024, 40(2): 266-276. |
| [3] | 潘虎, 周子琼, 田云. 三株异菌脲高效降解菌株的筛选、鉴定及其降解特性分析[J]. 生物技术通报, 2023, 39(6): 298-307. |
| [4] | 孙海航, 官会林, 王旭, 王童, 李泓霖, 彭文洁, 刘柏桢, 樊芳玲. 生物炭对三七连作土壤性质及真菌群落的影响[J]. 生物技术通报, 2023, 39(2): 221-231. |
| [5] | 王子夜, 王志刚, 阎爱华. 不同树龄桑根际土壤原生生物群落组成多样性[J]. 生物技术通报, 2022, 38(8): 206-215. |
| [6] | 陈天赐, 武少兰, 杨国辉, 江丹霞, 江玉姬, 陈炳智. 无柄灵芝醇提物对小鼠睡眠及肠道菌群的影响[J]. 生物技术通报, 2022, 38(8): 225-232. |
| [7] | 钟辉, 刘亚军, 王滨花, 和梦洁, 吴兰. 分析方法对细菌群落16S rRNA基因扩增测序分析结果的影响[J]. 生物技术通报, 2022, 38(6): 81-92. |
| [8] | 赵林艳, 官会林, 向萍, 李泽诚, 柏雨龙, 宋洪川, 孙世中, 徐武美. 白及根腐病植株根际土壤微生物群落组成特征分析[J]. 生物技术通报, 2022, 38(2): 67-74. |
| [9] | 陈宇捷, 郑华宝, 周昕彦. 改良高通量测序技术揭示除藻剂对藻类群落的影响[J]. 生物技术通报, 2022, 38(11): 70-79. |
| [10] | 颜珲璘, 芦光新, 邓晔, 顾松松, 颜程良, 马坤, 赵阳安, 张海娟, 王英成, 周学丽, 窦声云. 高寒地区根瘤菌拌种对禾/豆混播土壤微生物群落的影响[J]. 生物技术通报, 2022, 38(10): 204-215. |
| [11] | 曹修凯, 王珊, 葛玲, 张卫博, 孙伟. 染色体外环形DNA研究进展及其在畜禽育种中的应用[J]. 生物技术通报, 2022, 38(1): 247-257. |
| [12] | 康子清, 张银龙, 吴永波, 谢冬, 薛建辉, 华建峰. 环境DNA宏条形码在生物多样性研究与监测中的应用[J]. 生物技术通报, 2022, 38(1): 299-310. |
| [13] | 辛亚芬, 陈晨, 曾泰儒, 杜昭昌, 倪浩然, 钟怡豪, 谭小平, 闫艳红. 青贮添加剂对微生物多样性影响的研究进展[J]. 生物技术通报, 2021, 37(9): 24-30. |
| [14] | 王婷, 杨阳, 李金萍, 杜坤. 转基因作物对土壤微生物群落影响的研究进展[J]. 生物技术通报, 2021, 37(9): 255-265. |
| [15] | 张颖超, 尹守亮, 王一炜, 王学凯, 杨富裕. 木本饲料青贮研究进展[J]. 生物技术通报, 2021, 37(9): 48-57. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||