








生物技术通报 ›› 2024, Vol. 40 ›› Issue (12): 84-92.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0420
邹婷婷( ), 杨莉, 曲茜彤, 陈子航, 潘曦, 邵鸿旭, 王小丽(
), 杨莉, 曲茜彤, 陈子航, 潘曦, 邵鸿旭, 王小丽( )
)
收稿日期:2024-05-06
出版日期:2024-12-26
发布日期:2025-01-15
通讯作者:
王小丽,女,博士,副教授,研究方向:蔬菜栽培与品质调控;E-mail: wangxl@shnu.edu.cn作者简介:邹婷婷,女,硕士研究生,研究方向:蔬菜栽培与品质调控;E-mail: 2974567001@qq.com
基金资助:
ZOU Ting-ting( ), YANG Li, QU Xi-tong, CHEN Zi-hang, PAN Xi, SHAO Hong-xu, WANG Xiao-li(
), YANG Li, QU Xi-tong, CHEN Zi-hang, PAN Xi, SHAO Hong-xu, WANG Xiao-li( )
)
Received:2024-05-06
Published:2024-12-26
Online:2025-01-15
摘要:
【目的】高迁移率族蛋白(high mobility group, HMG)参与DNA结构/染色质的调节,在基因转录调控中起重要作用。鉴定菠菜(Spinacia oleracea L.)HMG基因家族,为菠菜HMG蛋白生物学功能研究提供理论依据。【方法】从全基因组水平对菠菜SoHMG家族成员进行鉴定,并对家族成员进行理化性质、基因结构、保守基序、启动子元件及水杨酸响应表达谱等分析。【结果】菠菜基因组含有16个SoHMG基因家族成员,编码区长度为240-4 029 bp,编码氨基酸79-1 342,分散在6条染色体上。进化树分析将SoHMG划分为2个亚家族,包括3个A亚族和13个B亚族成员,同一亚家族成员具有类似的保守结构域。在B亚家族启动子中发现了水杨酸等激素响应元件。SoHMG基因在菠菜根、茎、叶中均有表达,其中4个基因在叶中表达量较高。绝大多数SoHMG基因表达量在水杨酸处理下表达上调,其中,SoHMGB1(SOV1g000250.1)、SoHMGB7(SOV5g026260.1)、SoHMGB2(SOV1g027200.1)上调幅度最为显著,上调倍数10倍以上。【结论】菠菜HMG基因家族注释得到16个成员,分为2个亚族,其中3个基因(SoHMGB1、SoHMGB7和SoHMGB2)在水杨酸处理下显著上调,可能在水杨酸信号响应中发挥重要作用。
邹婷婷, 杨莉, 曲茜彤, 陈子航, 潘曦, 邵鸿旭, 王小丽. 菠菜高迁移率族基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(12): 84-92.
ZOU Ting-ting, YANG Li, QU Xi-tong, CHEN Zi-hang, PAN Xi, SHAO Hong-xu, WANG Xiao-li. Identification and Expression Analysis of High Mobility Gene Families in Spinach[J]. Biotechnology Bulletin, 2024, 40(12): 84-92.
| 基因编号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| SOV1g000250.1 | TGGTGAACTCTGGAACAAA | CTCGTCTTCATCAGTCTCG |
| SOV1g027200.1 | TCAACTACGGAGCGATGGC | TCCACATAGCAGAAACAGC |
| SOV2g035530.1 | GCAACGACCGTGGAAAAAC | CCTTGACACCCTTGGAATC |
| SOV3g013500.1 | GTTGCTATTACTGATGCTGC | AGGAATGTGATACAAGATGC |
| SOV4g023910.1 | AAAAGAGAACCCAAATGCAG | CGCGCTTTCTTTGCTGACA |
| SOV5g003140.1 | CTGCATCTTTCATTCTTACGTC | TTCAACAACATAGTCGGTCT |
| SOV5g026260.1 | CATCAAAGAGGTCAAAATCG | TATTCTGCCATTGCCCTGT |
| SOV5g034730.1 | TTTTATGCCTCGGGTATTAGCC | TGCCTGCTGCTTCATCTGA |
| SOV5g037650.1 | CCAAGTGCCTTCTTCGTCT | GCCATCTTCATCATCCTCA |
| SOV6g009370.1 | AAGACTCAGACTCACTCGG | GGGACTATTGCTAACCTAA |
| SOV6g009420.1 | CACACGAAAGAGTTGAAGA | GGGACTATTGCTAACCTAA |
| SOV6g043170.1 | AAGCCTACTATGACTCTGCC | CACTTTGCACCCATTATGTT |
| SOV4g034920.1 | CCCAACTTCATCCGTCGTG | AAACCCCTTCACCCCCAAT |
| SOV2g030330.1 | GCCTACTAAGGGGATTGTGTGGA | TCATTGGCGGTTCATCAGG |
| SOV4g003160.1 | TCTGTTGGGCTGGATTCGC | CTCTTCCTTGGACGACCTCT |
表1 实时荧光定量PCR引物序列
Table 1 Primer sequences for real-time fluorescence quantitative PCR
| 基因编号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| SOV1g000250.1 | TGGTGAACTCTGGAACAAA | CTCGTCTTCATCAGTCTCG |
| SOV1g027200.1 | TCAACTACGGAGCGATGGC | TCCACATAGCAGAAACAGC |
| SOV2g035530.1 | GCAACGACCGTGGAAAAAC | CCTTGACACCCTTGGAATC |
| SOV3g013500.1 | GTTGCTATTACTGATGCTGC | AGGAATGTGATACAAGATGC |
| SOV4g023910.1 | AAAAGAGAACCCAAATGCAG | CGCGCTTTCTTTGCTGACA |
| SOV5g003140.1 | CTGCATCTTTCATTCTTACGTC | TTCAACAACATAGTCGGTCT |
| SOV5g026260.1 | CATCAAAGAGGTCAAAATCG | TATTCTGCCATTGCCCTGT |
| SOV5g034730.1 | TTTTATGCCTCGGGTATTAGCC | TGCCTGCTGCTTCATCTGA |
| SOV5g037650.1 | CCAAGTGCCTTCTTCGTCT | GCCATCTTCATCATCCTCA |
| SOV6g009370.1 | AAGACTCAGACTCACTCGG | GGGACTATTGCTAACCTAA |
| SOV6g009420.1 | CACACGAAAGAGTTGAAGA | GGGACTATTGCTAACCTAA |
| SOV6g043170.1 | AAGCCTACTATGACTCTGCC | CACTTTGCACCCATTATGTT |
| SOV4g034920.1 | CCCAACTTCATCCGTCGTG | AAACCCCTTCACCCCCAAT |
| SOV2g030330.1 | GCCTACTAAGGGGATTGTGTGGA | TCATTGGCGGTTCATCAGG |
| SOV4g003160.1 | TCTGTTGGGCTGGATTCGC | CTCTTCCTTGGACGACCTCT |
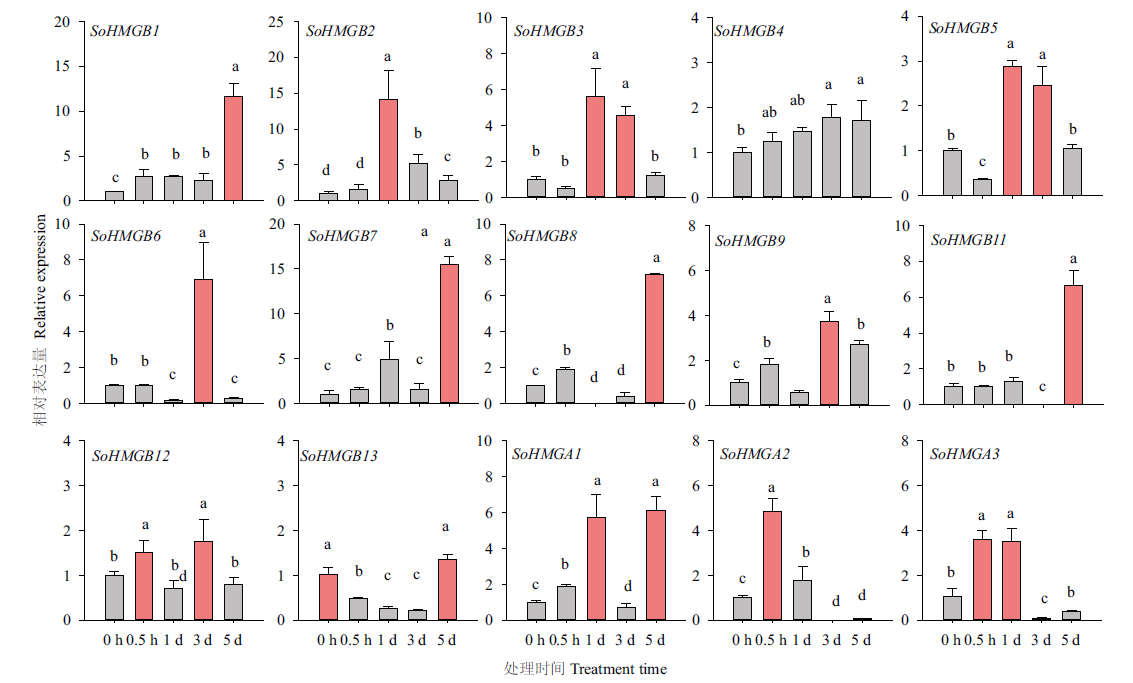
图7 菠菜水杨酸处理后SoHMG基因相对表达量 不同小写字母表示处理间差异显著(Tukey’test, P<0.05)
Fig. 7 Relative expressions of SoHMG gene in spinach after SA treatment Different lower-case letters indicate significant differences between treatments(Tukey’test, P<0.05)
| [1] | Cai XF, Sun XP, Xu CX, et al. Genomic analyses provide insights into spinach domestication and the genetic basis of agronomic traits[J]. Nat Commun, 2021, 12(1): 7246. |
| [2] | Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1[J]. Science, 1989, 243(4894Pt1): 1056-1059. |
| [3] | Grasser KD, Launholt D, Grasser M. High mobility group proteins of the plant HMGB family: Dynamic chromatin modulators[J]. Biochim Biophys Acta, 2007, 1769(5/6): 346-357. |
| [4] |
Antosch M, Mortensen SA, Grasser KD. Plant proteins containing high mobility group box DNA-binding domains modulate different nuclear processes[J]. Plant Physiol, 2012, 159(3): 875-883.
doi: 10.1104/pp.112.198283 pmid: 22585776 |
| [5] |
Hu LS, Yang XY, Yuan DJ, et al. GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton.[J]. Plant Biotechnol J, 2011, 9(9): 1038-1048.
doi: 10.1111/j.1467-7652.2011.00617.x pmid: 21554528 |
| [6] | Hamilton DJ, Hein AE, Wuttke DS, et al. The DNA binding high mobility group box protein family functionally binds RNA[J]. Wiley Interdiscip Rev RNA, 2023, 14(5): e1778. |
| [7] | Stemmer C, Schwander A, Bauw G, et al. Protein kinase CK2 differentially phosphorylates maize chromosomal high mobility group B(HMGB)proteins modulating their stability and DNA interactions[J]. J Biol Chem, 2002, 277(2): 1092-1098. |
| [8] |
王春瑶, 雷晓锦, 刘中原. 逆境胁迫下山新杨PdbHMGs基因表达模式分析[J]. 植物研究, 2023, 43(6): 932-942.
doi: 10.7525/j.issn.1673-5102.2023.06.015 |
| Wang CY, Lei XJ, Liu ZY. Expression pattern analysis of PdbHMGs genes in Populus davidiana × P. bolleana under abiotic stress[J]. Bull Bot Res, 2023, 43(6): 932-942. | |
| [9] | Kwak KJ, Kim JY, Kim YO, et al. Characterization of transgenic arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress[J]. Plant and Cell Physiol, 2007, 48(2): 221-231. |
| [10] | Villano C, D'Amelia V, Esposito S, et al. Genome-wide HMG family investigation and its role in glycoalkaloid accumulation in wild tuber-bearing Solanum commersonii[J]. Life, 2020, 10(4): 37. |
| [11] | Xu K, Chen SJ, Li TF, et al. Overexpression of OsHMGB707, a high mobility group protein, enhances rice drought tolerance by promoting stress-related gene expression[J]. Front Plant Sci, 2021, 12: 711271. |
| [12] |
Jang JY, Kwak KJ, Kang H. Expression of a high mobility group protein isolated from Cucumis sativus affects the germination of Arabidopsis thaliana under abiotic stress conditions[J]. J Integr Plant Biol, 2008, 50(5): 593-600.
doi: 10.1111/j.1744-7909.2008.00650.x |
| [13] | Choi HW, Manohar M, Manosalva P, et al. Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid[J]. PLoS Pathog, 2016, 12(3): e1005518. |
| [14] | 张小茜, 全先庆, 单雷, 等. 高迁移率族蛋白A的特征及其在转录中的调控[J]. 生命的化学, 2006, 26(5): 414-417. |
| Zhang XQ, Quan XQ, Shan L, et al. Characteristics of high mobility group protein A and its regulation in transcription[J]. Chem Life, 2006, 26(5): 414-417. | |
| [15] | Zhao J, Paul LK, Grafi G. The maize HMGA protein is localized to the nucleolus and can be acetylated in vitro at its globular domain, and phosphorylation by CDK reduces its binding activity to AT-rich DNA[J]. Biochim Biophys Acta, 2009, 1789(11/12): 751-757. |
| [16] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: A “One for All, All for One” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [17] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCtmethod[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [18] | Wang XL, Cai XF, Xu CX, et al. Identification and characterization of the NPF, NRT2 and NRT3in spinach[J]. Plant Physiol Biochem, 2021, 158: 297-307. |
| [19] |
Wu Q, Zhang WS, Pwee KH, et al. Cloning and characterization of rice HMGB1 gene[J]. Gene, 2003, 312: 103-109.
pmid: 12909345 |
| [20] |
Tang HB, Wang XY, Bowers JE, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps[J]. Genome Res, 2008, 18(12): 1944-1954.
doi: 10.1101/gr.080978.108 pmid: 18832442 |
| [21] | Luthringer R, Raphalen M, Guerra C, et al. Repeated co-option of HMG-box genes for sex determination in brown algae and animals[J]. Science, 2024, 383(6689): eadk5466. |
| [22] | Young Jang J, Jin Kwak K, Kang H. Molecular cloning of a cDNA encoding a high mobility group protein in Cucumis sativus and its axpression by abiotic stress treatments[J]. J Plant Physiol, 2007, 164(2): 205-208. |
| [23] | 时明坤. TaHMGB家族的生物信息学分析与TaHMGB11-7A功能的初步鉴定[D]. 郑州: 河南农业大学, 2021. |
| Shi MK. Bioinformatics analysis of TaHMGB family and preliminary functional identification of TaHMGB11-7A function[D]. Zhengzhou: Henan Agricultural University, 2021. | |
| [24] | Kaya C, Ugurlar F, Ashraf M, et al. Salicylic acid interacts with other plant growth regulators and signal molecules in response to stressful environments in plants[J]. Plant Physiol Biochem, 2023, 196: 431-443. |
| [25] | Ullah C, Schmidt A, Reichelt M, et al. Lack of antagonism between salicylic acid and jasmonate signalling pathways in poplar[J]. New phytol, 2022, 235(2): 701-717. |
| [26] | Rastegar S, Shojaie A, Koy RAM. Foliar application of salicylic acid and calcium chloride delays the loss of chlorophyll and preserves the quality of broccoli during storage[J]. J Food Biochem, 2022, 46(8): e14154. |
| [27] |
Puthusseri B, Divya P, Lokesh V, et al. Salicylic acid-induced elicitation of folates in coriander(Coriandrum sativum L.) improves bioaccessibility and reduces pro-oxidant status[J]. Food Chem, 2013, 136(2): 569-575.
doi: 10.1016/j.foodchem.2012.09.005 pmid: 23122099 |
| [28] | 王惠宇, 门艺涵, 樊婕, 等. 外源水杨酸影响苦荞类黄酮生物合成及相关基因表达模式[J]. 植物生理学报, 2024, 60(1): 117-129. |
| Wang HY, Men YH, Fan J, et al. Effects of exogenous salicylic acid on flavonoid biosynthesis and expression patterns of related genes in tartary buckwheat[J]. Plant Physiol J, 2024, 60(1): 117-129 | |
| [29] |
张云秀, 蒋旭, 尉春雪, 等. 紫花苜蓿高迁移率族蛋白基因MsHMG-Y调控花期的功能分析[J]. 中国农业科学, 2022, 55(16): 3082-3092.
doi: 10.3864/j.issn.0578-1752.2022.16.002 |
| Zhang YX, Jiang X, Yu CX, et al. The functional analysis of high mobility group MsHMG-Y involved in flowering regulation in Medicago sativa L[J]. Sci Agric Sin, 2022, 55(16): 3082-3092. | |
| [30] | Mollah MMI, Choi HW, Yeam I, et al. Salicylic acid, a plant hormone, suppresses phytophagous insect immune response by interrupting HMG-like DSP1[J]. Front Physiol, 2021, 12: 744272. |
| [1] | 何财林, 卢晶, 郭会会, 李小安, 吴琪. 藜麦MADS-box基因家族的全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(1): 157-172. |
| [2] | 刘倩, 马连杰, 张慧, 王冬, 范茂, 廖敦秀, 赵正武, 卢文才. 辣椒炭疽病生防菌株TN2的筛选鉴定与抑菌效果[J]. 生物技术通报, 2025, 41(1): 287-297. |
| [3] | 张亚亚, 李盼盼, 高惠惠, 贾晨波, 徐春燕. 基于根表真菌群落与病原菌鉴定探究‘宁杞5号’枸杞根腐病的发生机制[J]. 生物技术通报, 2024, 40(9): 238-248. |
| [4] | 袁兰, 黄娅楠, 张贝妮, 熊雨萌, 王洪洋. 基于流式细胞仪鉴定马铃薯倍性的高通量样品制备方法[J]. 生物技术通报, 2024, 40(9): 141-147. |
| [5] | 杜薇, 李志敏, 邢晏铭, 刘蒲临, 缪礼鸿. 一株易转化、高生物量地衣芽孢杆菌的筛选与鉴定[J]. 生物技术通报, 2024, 40(9): 181-189. |
| [6] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [7] | 谭博文, 张懿, 张鹏, 王振宇, 马秋香. 木薯镁离子转运蛋白家族基因的鉴定及生物信息学分析[J]. 生物技术通报, 2024, 40(9): 20-32. |
| [8] | 张曼玉, 董嘉诚, 苟福凡, 弓朝晖, 刘倩, 孙文良, 孔臻, 郝捷, 王敏, 田朝光. 嗜热毁丝霉果胶酯酶MtCE12-1的克隆表达及其酶学性质和应用研究[J]. 生物技术通报, 2024, 40(9): 291-300. |
| [9] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [10] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [11] | 王茜, 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建. 向日葵YABBY基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(8): 199-211. |
| [12] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [13] | 王芳, 于璐, 齐泽铮, 周长军, 于吉东. 大豆镰刀菌根腐病拮抗菌的筛选及生防效果[J]. 生物技术通报, 2024, 40(7): 216-225. |
| [14] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [15] | 杨鹭, 袁源, 方志锴, 林如, 江红, 周剑. 一株链霉菌的鉴定及其产格尔德霉素的发酵工艺研究[J]. 生物技术通报, 2024, 40(6): 299-309. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||