








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 255-270.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0618
• 研究报告 • 上一篇
收稿日期:2024-07-01
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
肖艳辉,女,教授,研究方向 :芳香植物栽培生理与利用;E-mail: xiaoyanhui-7394@163.com作者简介:王斌,男,博士,副教授,研究方向 :园艺植物逆境生物学;E-mail: b_wang@sgu.edu.cn
基金资助:
WANG Bin( ), WANG Yu-kun, XIAO Yan-hui(
), WANG Yu-kun, XIAO Yan-hui( )
)
Received:2024-07-01
Published:2025-03-26
Online:2025-03-20
摘要:
目的 丁香罗勒(Ocimum gratissimum)同时具有修复和安全利用镉(cadmium, Cd)污染土壤的潜力,解析丁香罗勒响应Cd胁迫的分子机制,鉴定与Cd耐受性密切相关的调控因子,为耐受Cd胁迫的丁香罗勒种质资源创新提供候选基因资源。 方法 采用比较转录组学的方法,分析Cd处理(35 μmol/L)72 h期间丁香罗勒叶片的转录组学变化。 结果 Cd胁迫显著抑制丁香罗勒生长,叶片、茎和根中均积累了很高含量的Cd,根中的Cd含量最高。Cd胁迫还显著影响精油组分和含量,Cd处理诱导肉桂酸甲酯的合成积累。转录组分析结果显示,在24 h的比较组中,差异表达基因(differentially expressed genes, DEGs)富集在核糖体、植物-病原相互作用和MAPK信号通路等途径中。在72 h的比较组中,DEGs富集在光合作用、光合作用-天线蛋白和卟啉与叶绿素代谢途径中。浅青色1模块内的基因与Cd胁迫表型显著相关,模块内的基因主要富集在植物激素信号转导、植物-病原相互作用和MAPK信号通路途径中。Cd胁迫上调表达的DEGs也显著富集在这3个途径,表明激活这3个途径是丁香罗勒叶片应答Cd胁迫的主要机制。Cd胁迫处理诱导13个转录因子基因表达上调,其中5个是bHLH家族基因,表明bHLH转录因子在丁香罗勒Cd胁迫耐受性调控中具有重要作用。 结论 丁香罗勒应答Cd胁迫的分子机制与其他植物有很大相似之处,bHLH转录因子可能是调控丁香罗勒Cd耐受性的关键转录因子。
王斌, 王玉昆, 肖艳辉. 丁香罗勒(Ocimum gratissimum)叶片响应镉胁迫的比较转录组学分析[J]. 生物技术通报, 2025, 41(3): 255-270.
WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress[J]. Biotechnology Bulletin, 2025, 41(3): 255-270.
基因编号 Gene ID | 正向引物序列 Forward primer sequence (5΄‒3΄) | 反向引物序列 Reverse primer sequence (5΄‒3΄) | 产物长度 Product length/bp |
|---|---|---|---|
| c52549.graph_c0 | ATCGAGCAGAGATGAAGCCG | TTGACGCGCGTGTTTTTCAT | 134 |
| c49765.graph_c0 | AACAAGAGGGCGGAAGGAAG | AGAGAAAGAGGGGCTGTTGC | 73 |
| c57224.graph_c0 | AAGGCGCTGGAATCGGATAG | GCGATTTGCACTCCACGTTT | 125 |
| c59334.graph_c0 | CACAATTCCGATCGCACGAC | TGGCCTCCAAGTTATGCTGG | 71 |
| c51522.graph_c0 | CGGTTCGTCTCCTTGGTGAA | AGTCTCCTCGACCAGCTCAT | 119 |
| c54882.graph_c1 | AGGATGGTGTGAGGAGAGCA | TGCTCAAGTGGAGGATGCAA | 140 |
| Actin7 | GGAGCTCGTCTTTGCTGTCT | GAGCGGGAAATTGTGAGGGA | 90 |
表1 本试验所用引物
Table 1 Primers used in this study
基因编号 Gene ID | 正向引物序列 Forward primer sequence (5΄‒3΄) | 反向引物序列 Reverse primer sequence (5΄‒3΄) | 产物长度 Product length/bp |
|---|---|---|---|
| c52549.graph_c0 | ATCGAGCAGAGATGAAGCCG | TTGACGCGCGTGTTTTTCAT | 134 |
| c49765.graph_c0 | AACAAGAGGGCGGAAGGAAG | AGAGAAAGAGGGGCTGTTGC | 73 |
| c57224.graph_c0 | AAGGCGCTGGAATCGGATAG | GCGATTTGCACTCCACGTTT | 125 |
| c59334.graph_c0 | CACAATTCCGATCGCACGAC | TGGCCTCCAAGTTATGCTGG | 71 |
| c51522.graph_c0 | CGGTTCGTCTCCTTGGTGAA | AGTCTCCTCGACCAGCTCAT | 119 |
| c54882.graph_c1 | AGGATGGTGTGAGGAGAGCA | TGCTCAAGTGGAGGATGCAA | 140 |
| Actin7 | GGAGCTCGTCTTTGCTGTCT | GAGCGGGAAATTGTGAGGGA | 90 |

图1 Cd胁迫处理对丁香罗勒生长的影响A:株高;B:叶片相对叶绿素含量;C:地上部鲜重;D:地上部干重;E:叶宽;F:叶长;G:叶片表型;H:整株表型;I:根鲜重;J:根干重;K:根系轮廓。不同小写字母表示在P<0.05水平上差异显著。下同
Fig. 1 Effects of Cd stress treatment on the growth of clove basilA: Plant height. B: SPAD. C: Fresh weight of aboveground parts. D: Dry weight of aboveground parts. E: Leaf width. F: Leaf length. G: Leaf phenotype. H: Whole plant phenotype. I: Fresh weight of root. J: Dry weight of root. K: Root profile. Different lowercase letters indicate significant differences at P<0.05 level. The same below
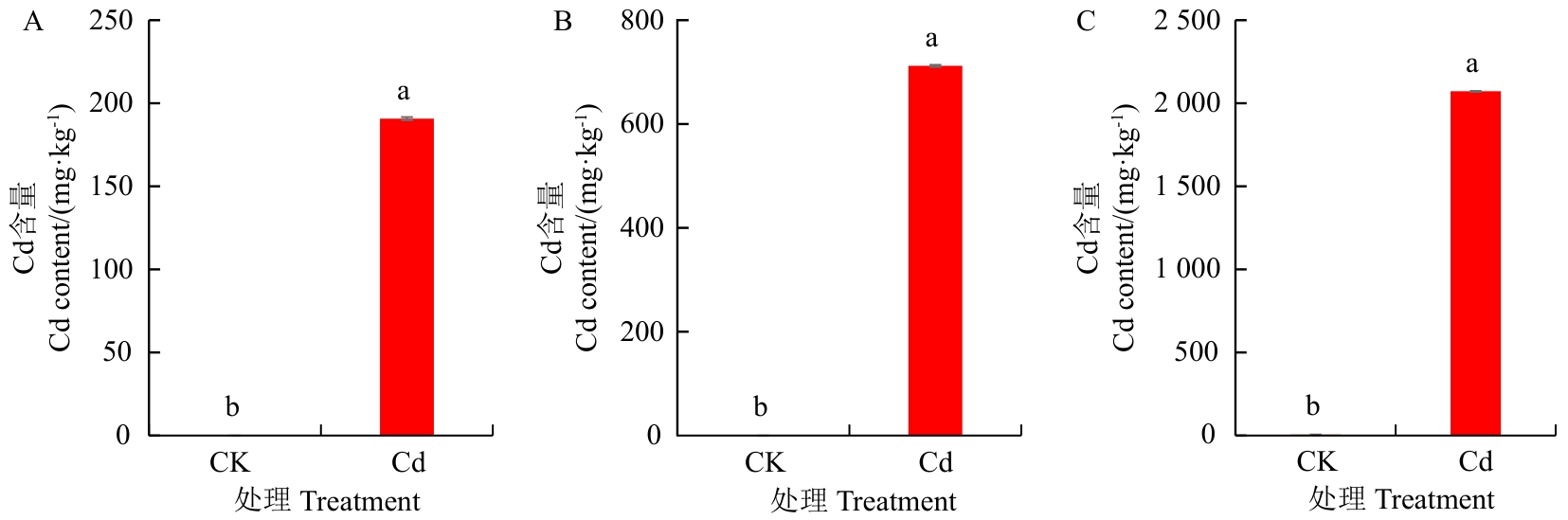
图2 丁香罗勒各组织中的Cd含量A:叶片中的Cd含量;B:茎中的Cd含量;C:根中的Cd含量
Fig. 2 Cd contents in varying organisms of clove basilA: Cd content in the leaves. B: Cd content in the shoots. C: Cd content in the roots
序号 No. | 化合物名称 Compound name | 保留时间 Retention time/min | 含量 Content/% | 序号 No. | 化合物名称 Compound name | 保留时间 Retention time/min | 含量 Content/% | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CK | Cd | CK | Cd | ||||||||
| 1 | 3-己烯醛 3-hexenal | 4.13 | 1.86±0.16* | 0.68±0.04 | 26 | 顺式-肉桂酸甲酯 Methyl cis-cinnamate | 12.00 | - | 5.13±0.26 | ||
| 2 | (E)-2-己烯醛 (E)-2-hexenal | 4.96 | 0.25±0.01* | 0.08±0.00 | 27 | 丁香酚Eugenol | 12.71 | 25.7±1.41* | 0.02±0.01 | ||
| 3 | 顺式-3-己烯醇 Cis-3-hexenol | 5.00 | 1.54±0.41* | 0.14±0.04 | 28 | 肉桂酸甲酯 Methyl ester cinnamic acid | 13.06 | 0.06±0.10 | 58.02±2.02* | ||
| 4 | α-侧柏烯 α-thujene | 6.16 | 0.15±0.02* | 0.06±0.01 | 29 | β-榄香烯 β-elemene | 13.21 | 0.83±0.09* | 0.31±0.04 | ||
| 5 | α-蒎烯 α-pinene | 6.28 | 0.38±0.02* | 0.21±0.01 | 30 | 甲基丁香酚 Methyl eugenol | 13.28 | 6.56±0.14* | 2.04±0.31 | ||
| 6 | 莰烯 Camphene | 6.54 | 0.06±0.01 | 0.02±0.00 | 31 | β-佛手柑油烯 β-bergamotene | 13.49 | 0.10±0.01 | 0.05±0.01 | ||
| 7 | 香桧烯 Sabinene | 6.93 | 0.14±0.01* | 0.08±0.01 | 32 | 石竹烯 Caryophyllene | 13.64 | 0.31±0.02* | 0.16±0.01 | ||
| 8 | β-蒎烯β-pinene | 7.00 | 0.38±0.04* | 0.19±0.01 | 33 | 反式-α-佛手柑油烯 Trans-α-bergamotene | 13.76 | 2.41±0.11 | 2.09±0.05 | ||
| 9 | β-月桂烯 β-myrcene | 7.18 | 0.02±0.01 | 0.10±0.00* | 34 | 顺式-β-金合欢烯Cis-β-farnesene | 13.83 | 0.21±0.02 | 0.08±0.02 | ||
| 10 | D-柠檬烯 D-limonene | 7.83 | 0.32±0.00* | 0.15±0.00 | 35 | 反式-β-金合欢烯 Trans-β-farnesene | 13.95 | 0.29±0.03* | 0.19±0.02 | ||
| 11 | 桉树脑 Eucalyptol | 7.89 | 3.02±0.11* | 1.95±0.09 | 36 | 蛇麻烯 Humulene | 14.08 | 1.27±0.04* | 0.70±0.02 | ||
| 12 | (E)-β-罗勒烯 (E)-β-ocimene | 8.10 | 2.27±0.11* | 1.13±0.02 | 37 | 顺式-4(15),5-依兰油二烯 Cis-muurola-4(15),5-diene | 14.18 | 0.25±0.05* | 0.08±0.01 | ||
| 13 | γ-松油烯 γ-terpinene | 8.30 | 0.40±0.02* | 0.14±0.01 | 38 | 大根香叶烯 D Germacrene D | 14.41 | 2.52±0.04* | 1.18±0.02 | ||
| 14 | 顺式-水合香桧烯Cis-sabinene hydrate | 8.46 | 0.79±0.05* | 0.27±0.01 | 39 | β-环大根香叶烷 β-cyclogermacrane | 14.61 | 1.54±0.09* | 0.4±0.01 | ||
| 15 | 萜品油烯 Terpinolene | 8.78 | 0.14±0.01* | 0.01±0.02 | 40 | δ-愈创木烯 δ-guaiene | 14.69 | 0.21±0.02* | 0.06±0.02 | ||
| 16 | 葑酮 Fenchone | 8.82 | 0.23±0.02 | 0.23±0.05 | 41 | γ-杜松烯 γ-cadinene | 14.80 | 1.07±0.04* | 0.56±0.02 | ||
| 17 | 芳樟醇 Linalool | 8.92 | 12.10±0.51* | 4.15±0.19 | 42 | β-倍半水芹烯 β-sesquiphellandrene | 14.86 | 0.11±0.03 | 0.06±0.01 | ||
| 18 | (+)-2-莰酮 (+)-2-bornanone | 9.71 | 0.85±0.03* | 0.25±0.01 | 43 | α-杜松烯 α-cadinene | 15.01 | 0.09±0.02 | 0.03±0.00 | ||
| 19 | 新薄荷醇 Neo-menthol | 9.97 | 0.12±0.01* | 0.03±0.00 | 44 | 反式-橙花叔醇 Trans-nerolidol | 15.28 | 0.41±0.21* | 0.06±0.00 | ||
| 20 | δ-松油醇 δ-terpineol | 10.02 | 0.09±0.01 | 0.04±0.00 | 45 | 表荜澄茄油烯醇 Epicubenol | 16.03 | 0.20±0.02* | 0.08±0.01 | ||
| 21 | 4-萜品醇 L-terpinen-4-ol | 10.17 | 4.46±0.76* | 2.65±0.12 | 46 | τ-杜松醇 τ-cadinol | 16.31 | 2.36±0.04* | 0.98±0.02 | ||
| 22 | L-α-松油醇 L-α-terpineol | 10.36 | 1.35±0.10* | 0.68±0.02 | 47 | 吲哚-3-乙醛 Indole-3-acetaldehyde | 16.96 | 0.08±0.03 | - | ||
| 23 | 4-烯丙基苯甲醚 Estragole | 10.46 | 20.50±1.47* | 13.9±2.26 | 48 | 新植二烯 Neophytadiene | 18.29 | 0.69±0.10* | 0.31±0.03 | ||
| 24 | 乙酸辛酯 Octyl ester acetic acid | 10.57 | 0.32±0.04* | 0.16±0.03 | 49 | n-十六酸 n-hexadecanoic acid | 19.46 | 0.18±0.01* | 0.08±0.00 | ||
| 25 | 3-乙烯基-4-甲基-1H- 吡咯-2,5-二酮 3-ethenyl-4-methyl-1H-pyrrole-2,5-dione | 11.25 | 0.17±0.01* | 0.05±0.00 | 合计 Total amount/% | 99.67 | 99.91 | ||||
表2 丁香罗勒叶片中的挥发性化合物及含量
Table 2 Volatile compounds and their contents in clove basil leaves
序号 No. | 化合物名称 Compound name | 保留时间 Retention time/min | 含量 Content/% | 序号 No. | 化合物名称 Compound name | 保留时间 Retention time/min | 含量 Content/% | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CK | Cd | CK | Cd | ||||||||
| 1 | 3-己烯醛 3-hexenal | 4.13 | 1.86±0.16* | 0.68±0.04 | 26 | 顺式-肉桂酸甲酯 Methyl cis-cinnamate | 12.00 | - | 5.13±0.26 | ||
| 2 | (E)-2-己烯醛 (E)-2-hexenal | 4.96 | 0.25±0.01* | 0.08±0.00 | 27 | 丁香酚Eugenol | 12.71 | 25.7±1.41* | 0.02±0.01 | ||
| 3 | 顺式-3-己烯醇 Cis-3-hexenol | 5.00 | 1.54±0.41* | 0.14±0.04 | 28 | 肉桂酸甲酯 Methyl ester cinnamic acid | 13.06 | 0.06±0.10 | 58.02±2.02* | ||
| 4 | α-侧柏烯 α-thujene | 6.16 | 0.15±0.02* | 0.06±0.01 | 29 | β-榄香烯 β-elemene | 13.21 | 0.83±0.09* | 0.31±0.04 | ||
| 5 | α-蒎烯 α-pinene | 6.28 | 0.38±0.02* | 0.21±0.01 | 30 | 甲基丁香酚 Methyl eugenol | 13.28 | 6.56±0.14* | 2.04±0.31 | ||
| 6 | 莰烯 Camphene | 6.54 | 0.06±0.01 | 0.02±0.00 | 31 | β-佛手柑油烯 β-bergamotene | 13.49 | 0.10±0.01 | 0.05±0.01 | ||
| 7 | 香桧烯 Sabinene | 6.93 | 0.14±0.01* | 0.08±0.01 | 32 | 石竹烯 Caryophyllene | 13.64 | 0.31±0.02* | 0.16±0.01 | ||
| 8 | β-蒎烯β-pinene | 7.00 | 0.38±0.04* | 0.19±0.01 | 33 | 反式-α-佛手柑油烯 Trans-α-bergamotene | 13.76 | 2.41±0.11 | 2.09±0.05 | ||
| 9 | β-月桂烯 β-myrcene | 7.18 | 0.02±0.01 | 0.10±0.00* | 34 | 顺式-β-金合欢烯Cis-β-farnesene | 13.83 | 0.21±0.02 | 0.08±0.02 | ||
| 10 | D-柠檬烯 D-limonene | 7.83 | 0.32±0.00* | 0.15±0.00 | 35 | 反式-β-金合欢烯 Trans-β-farnesene | 13.95 | 0.29±0.03* | 0.19±0.02 | ||
| 11 | 桉树脑 Eucalyptol | 7.89 | 3.02±0.11* | 1.95±0.09 | 36 | 蛇麻烯 Humulene | 14.08 | 1.27±0.04* | 0.70±0.02 | ||
| 12 | (E)-β-罗勒烯 (E)-β-ocimene | 8.10 | 2.27±0.11* | 1.13±0.02 | 37 | 顺式-4(15),5-依兰油二烯 Cis-muurola-4(15),5-diene | 14.18 | 0.25±0.05* | 0.08±0.01 | ||
| 13 | γ-松油烯 γ-terpinene | 8.30 | 0.40±0.02* | 0.14±0.01 | 38 | 大根香叶烯 D Germacrene D | 14.41 | 2.52±0.04* | 1.18±0.02 | ||
| 14 | 顺式-水合香桧烯Cis-sabinene hydrate | 8.46 | 0.79±0.05* | 0.27±0.01 | 39 | β-环大根香叶烷 β-cyclogermacrane | 14.61 | 1.54±0.09* | 0.4±0.01 | ||
| 15 | 萜品油烯 Terpinolene | 8.78 | 0.14±0.01* | 0.01±0.02 | 40 | δ-愈创木烯 δ-guaiene | 14.69 | 0.21±0.02* | 0.06±0.02 | ||
| 16 | 葑酮 Fenchone | 8.82 | 0.23±0.02 | 0.23±0.05 | 41 | γ-杜松烯 γ-cadinene | 14.80 | 1.07±0.04* | 0.56±0.02 | ||
| 17 | 芳樟醇 Linalool | 8.92 | 12.10±0.51* | 4.15±0.19 | 42 | β-倍半水芹烯 β-sesquiphellandrene | 14.86 | 0.11±0.03 | 0.06±0.01 | ||
| 18 | (+)-2-莰酮 (+)-2-bornanone | 9.71 | 0.85±0.03* | 0.25±0.01 | 43 | α-杜松烯 α-cadinene | 15.01 | 0.09±0.02 | 0.03±0.00 | ||
| 19 | 新薄荷醇 Neo-menthol | 9.97 | 0.12±0.01* | 0.03±0.00 | 44 | 反式-橙花叔醇 Trans-nerolidol | 15.28 | 0.41±0.21* | 0.06±0.00 | ||
| 20 | δ-松油醇 δ-terpineol | 10.02 | 0.09±0.01 | 0.04±0.00 | 45 | 表荜澄茄油烯醇 Epicubenol | 16.03 | 0.20±0.02* | 0.08±0.01 | ||
| 21 | 4-萜品醇 L-terpinen-4-ol | 10.17 | 4.46±0.76* | 2.65±0.12 | 46 | τ-杜松醇 τ-cadinol | 16.31 | 2.36±0.04* | 0.98±0.02 | ||
| 22 | L-α-松油醇 L-α-terpineol | 10.36 | 1.35±0.10* | 0.68±0.02 | 47 | 吲哚-3-乙醛 Indole-3-acetaldehyde | 16.96 | 0.08±0.03 | - | ||
| 23 | 4-烯丙基苯甲醚 Estragole | 10.46 | 20.50±1.47* | 13.9±2.26 | 48 | 新植二烯 Neophytadiene | 18.29 | 0.69±0.10* | 0.31±0.03 | ||
| 24 | 乙酸辛酯 Octyl ester acetic acid | 10.57 | 0.32±0.04* | 0.16±0.03 | 49 | n-十六酸 n-hexadecanoic acid | 19.46 | 0.18±0.01* | 0.08±0.00 | ||
| 25 | 3-乙烯基-4-甲基-1H- 吡咯-2,5-二酮 3-ethenyl-4-methyl-1H-pyrrole-2,5-dione | 11.25 | 0.17±0.01* | 0.05±0.00 | 合计 Total amount/% | 99.67 | 99.91 | ||||
样品编号 Sample No. | Clean reads | GC含量 GC content/% | ≥Q30/% | 比对读数 Mapped reads | 比对比率 Mapped ratio/% |
|---|---|---|---|---|---|
| 0 h CK-1 | 22 830 057 | 49.84 | 93.02 | 17 200 093 | 75.34 |
| 0 h CK-2 | 22 391 222 | 49.80 | 92.93 | 17 070 648 | 76.24 |
| 0 h CK-3 | 21 181 042 | 50.05 | 93.88 | 16 240 183 | 76.67 |
| 24 h CK-1 | 21 485 915 | 50.28 | 93.29 | 16 070 383 | 74.79 |
| 24 hCK-2 | 21 898 153 | 50.27 | 93.37 | 16 540 511 | 75.53 |
| 24 h CK-3 | 21 872 539 | 50.81 | 93.37 | 16 650 456 | 76.12 |
| 72 h CK-1 | 22 127 839 | 49.96 | 93.08 | 16 819 392 | 76.01 |
| 72 h CK-2 | 20 937 711 | 50.20 | 93.83 | 15 897 564 | 75.93 |
| 72 h CK-3 | 19 962 929 | 50.29 | 93.46 | 15 148 810 | 75.88 |
| 24 h Cd-1 | 25 353 059 | 49.97 | 93.80 | 19 449 566 | 76.71 |
| 24 h Cd-2 | 20 283 780 | 49.74 | 93.89 | 15 626 179 | 77.04 |
| 24 h Cd-3 | 20 638 087 | 49.93 | 93.68 | 15 794 277 | 76.53 |
| 72 h Cd-1 | 20 774 226 | 49.12 | 93.32 | 15 927 385 | 76.67 |
| 72 h Cd-2 | 26 674 376 | 48.82 | 92.73 | 19 912 643 | 74.65 |
| 72 h Cd-3 | 22 262 126 | 48.62 | 93.14 | 16 647 781 | 74.78 |
表3 测序数据评估与组装结果的比对
Table 3 Evaluation and comparison of sequencing data and assembly results
样品编号 Sample No. | Clean reads | GC含量 GC content/% | ≥Q30/% | 比对读数 Mapped reads | 比对比率 Mapped ratio/% |
|---|---|---|---|---|---|
| 0 h CK-1 | 22 830 057 | 49.84 | 93.02 | 17 200 093 | 75.34 |
| 0 h CK-2 | 22 391 222 | 49.80 | 92.93 | 17 070 648 | 76.24 |
| 0 h CK-3 | 21 181 042 | 50.05 | 93.88 | 16 240 183 | 76.67 |
| 24 h CK-1 | 21 485 915 | 50.28 | 93.29 | 16 070 383 | 74.79 |
| 24 hCK-2 | 21 898 153 | 50.27 | 93.37 | 16 540 511 | 75.53 |
| 24 h CK-3 | 21 872 539 | 50.81 | 93.37 | 16 650 456 | 76.12 |
| 72 h CK-1 | 22 127 839 | 49.96 | 93.08 | 16 819 392 | 76.01 |
| 72 h CK-2 | 20 937 711 | 50.20 | 93.83 | 15 897 564 | 75.93 |
| 72 h CK-3 | 19 962 929 | 50.29 | 93.46 | 15 148 810 | 75.88 |
| 24 h Cd-1 | 25 353 059 | 49.97 | 93.80 | 19 449 566 | 76.71 |
| 24 h Cd-2 | 20 283 780 | 49.74 | 93.89 | 15 626 179 | 77.04 |
| 24 h Cd-3 | 20 638 087 | 49.93 | 93.68 | 15 794 277 | 76.53 |
| 72 h Cd-1 | 20 774 226 | 49.12 | 93.32 | 15 927 385 | 76.67 |
| 72 h Cd-2 | 26 674 376 | 48.82 | 92.73 | 19 912 643 | 74.65 |
| 72 h Cd-3 | 22 262 126 | 48.62 | 93.14 | 16 647 781 | 74.78 |
数据库 Database | 注释的 基因数量 Annotated number | 长度在300‒1 000 bp的基因数量 300≤length<1 000 bp | 长度大于1 000 bp的基因数量 Length≥1 000 bp |
|---|---|---|---|
| COG | 9 606 | 2 703 | 5 395 |
| GO | 33 653 | 11 607 | 14 274 |
| KEGG | 26 168 | 8 557 | 12 005 |
| KOG | 21 510 | 6 891 | 9 885 |
| Pfam | 26 052 | 8 169 | 13 479 |
| Swissprot | 26 681 | 8 889 | 12 403 |
| TrEMBL | 39 794 | 14 047 | 16 646 |
| eggnog | 33 797 | 11 675 | 14 801 |
| NR | 40 191 | 14 083 | 16 642 |
注释的基因总数 All annotated genes | 41 060 | 14 425 | 16 718 |
表4 Unigene注释统计表
Table 4 Statistics table of unigene annotation
数据库 Database | 注释的 基因数量 Annotated number | 长度在300‒1 000 bp的基因数量 300≤length<1 000 bp | 长度大于1 000 bp的基因数量 Length≥1 000 bp |
|---|---|---|---|
| COG | 9 606 | 2 703 | 5 395 |
| GO | 33 653 | 11 607 | 14 274 |
| KEGG | 26 168 | 8 557 | 12 005 |
| KOG | 21 510 | 6 891 | 9 885 |
| Pfam | 26 052 | 8 169 | 13 479 |
| Swissprot | 26 681 | 8 889 | 12 403 |
| TrEMBL | 39 794 | 14 047 | 16 646 |
| eggnog | 33 797 | 11 675 | 14 801 |
| NR | 40 191 | 14 083 | 16 642 |
注释的基因总数 All annotated genes | 41 060 | 14 425 | 16 718 |
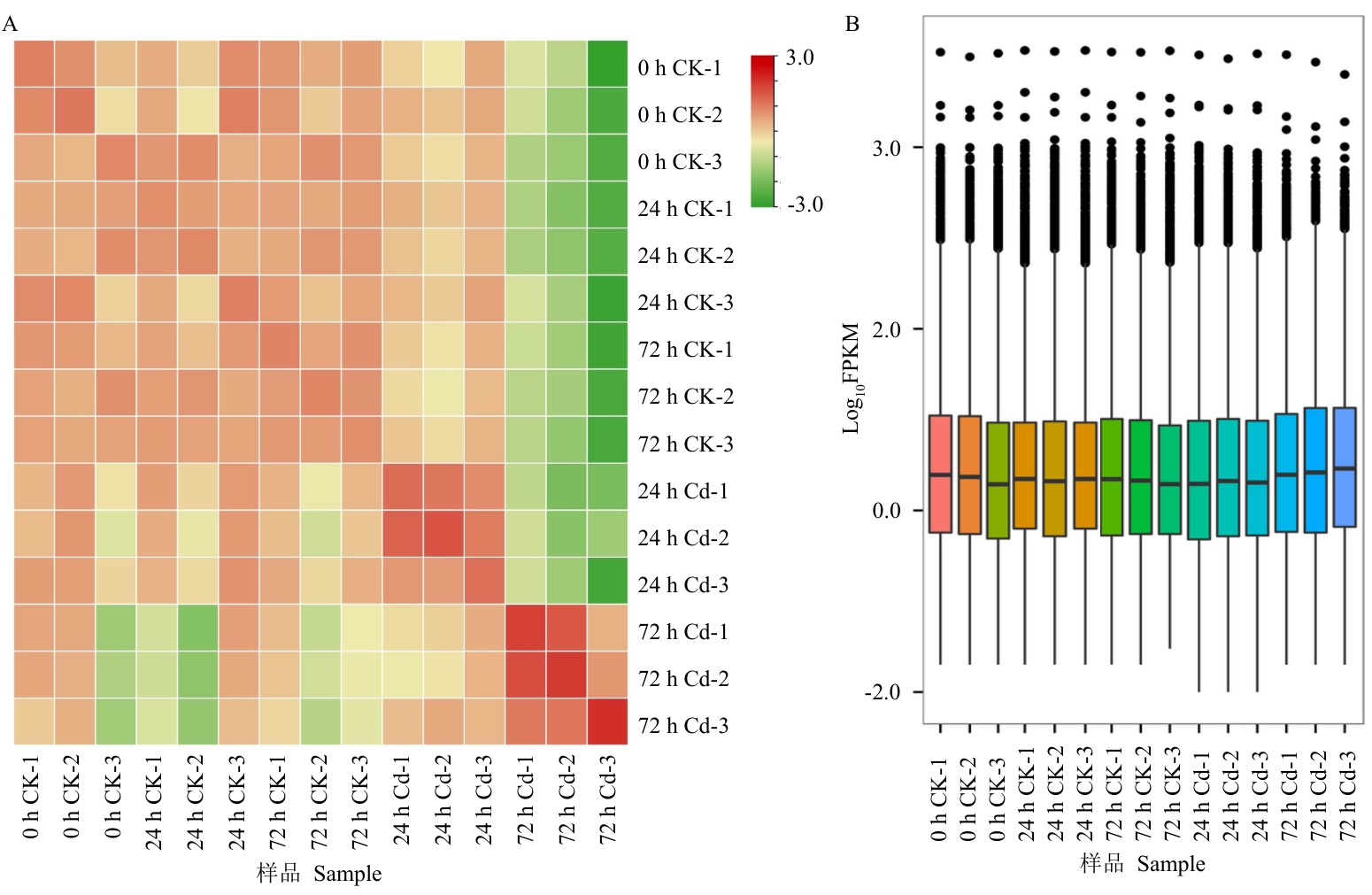
图3 样品相关性分析和整体基因表达A:样品相关性热图,每个色块表示一个样品;B:不同样品的整体表达丰度
Fig. 3 Sample correlation analysis and overall gene expressionA: Heat map of correlation analysis for varying samples. B: Overall gene expression abundance of varying samples

图5 Cd胁迫处理24 h的比较转录组分析A:火山图;B:所有差异表达基因的KEGG富集分析;C:上调表达的差异表达基因的KEGG富集分析;D:下调表达的差异表达基因的KEGG富集分析,下同
Fig. 5 Comparative transcriptomic analysis of Cd stress treatment at 24 hA: Volcano plot. B: KEGG enrichment analysis of all differentially expressed genes (DEGs). C: KEGG enrichment analysis of upregulated DEGs. D: KEGG enrichment analysis of downregulated DEGs, the same below

图7 加权基因共表达网络分析A:代表性基因模块的聚类树状图;B:模块与性状间的相关性分析;C:lightcyan1模块中的基因表达热图;D:lightcyan1模块中的基因表达与模块间的相关性;E:lightcyan1模块中的基因KEGG富集分析
Fig. 7 Weighted gene co-expression network analysisA: Dendrogram of clustered representative gene modules. B: Correlation analysis between modules and traits. C: Heatmap of gene expression in the lightcyan1 module. D: Correlation of module and gene expression within the lightcyan1.E: KEGG enrichment analysis of genes in the lightcyan1 module
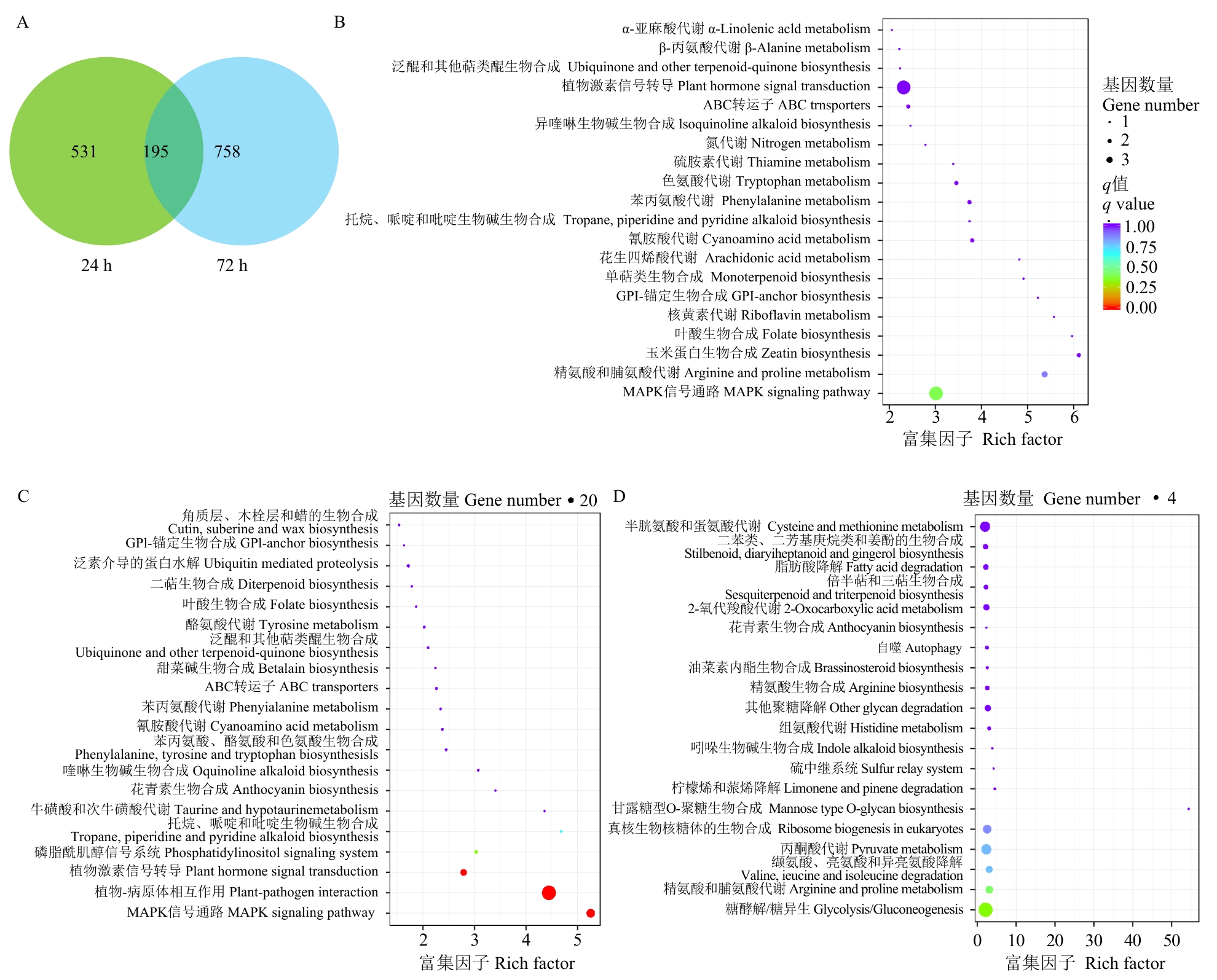
图8 Cd胁迫诱导的差异表达基因功能分析A:韦恩图;B:同时在24和72 h表达上调的差异表达基因的KEGG富集分析;C:仅在24 h上调表达的差异表达基因的KEGG富集分析;D:仅在72 h上调表达的差异表达基因的KEGG富集分析
Fig. 8 Functional analysis of differentially expressed genes induced by Cd stressA: Venn diagram. B: KEGG enrichment analysis of differentially expressed genes (DEGs) upregulated at both 24 h and 72 h. C: KEGG enrichment analysis of DEGs upregulated only at 24 h. D: KEGG enrichment analysis of DEGs upregulated solely at 72 h
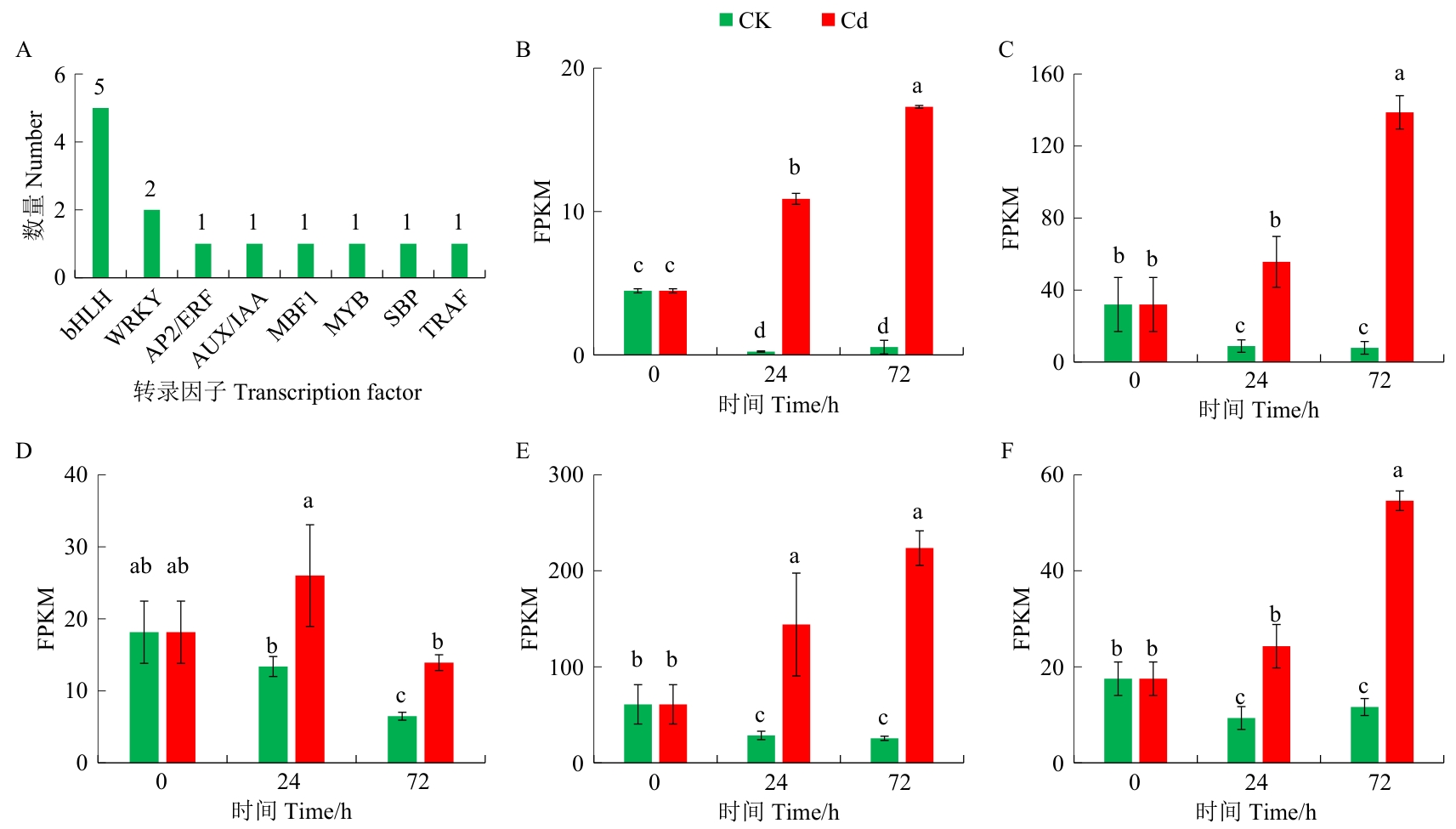
图9 Cd诱导的转录因子及5个bHLH基因在Cd胁迫处理期间的表达模式A:转录因子类型;B‒F:5个被Cd胁迫诱导的bHLH基因(基因ID依次为c51250.graph_c0、c51522.graph_c0、c54831.graph_c0、c56571.graph_c1和c57229.graph_c0)
Fig. 9 Transcription factors induced by Cd stress and the expression patterns of five bHLH genes during Cd stress durationA: Transcription factor types. B-F: 5 bHLH genes induced by Cd stress (gene IDs orderly is c51250.graph_c0, c51522.graph_c0, c54831.graph_c0, c56571.graph_c1, and c57229.graph_c0)
| 1 | 綦峥, 齐越, 杨红, 等. 土壤重金属镉污染现状、危害及治理措施 [J]. 食品安全质量检测学报, 2020, 11(7): 2286-2294. |
| Qi Z, Qi Y, Yang H, et al. Status, harm and treatment measures of heavy metal cadmium pollution in soil [J]. J Food Saf Qual, 2020, 11(7): 2286-2294. | |
| 2 | Satarug S, Baker JR, Urbenjapol S, et al. A global perspective on cadmium pollution and toxicity in non-occupationally exposed population [J]. Toxicol Lett, 2003, 137(1/2): 65-83. |
| 3 | 陈能场, 郑煜基, 何晓峰, 等. 《全国土壤污染状况调查公报》探析 [J]. 农业环境科学学报, 2017, 36(9): 1689-1692. |
| Chen NC, Zheng YJ, He XF, et al. Analysis of the report on the national general survey of soil contamination [J]. J Agro Environ Sci, 2017, 36(9): 1689-1692. | |
| 4 | Gavrilescu M. Enhancing phytoremediation of soils polluted with heavy metals [J]. Curr Opin Biotechnol, 2022, 74: 21-31. |
| 5 | Wang B, Lin LN, Yuan X, et al. Low-level cadmium exposure induced hormesis in peppermint young plant by constantly activating antioxidant activity based on physiological and transcriptomic analyses [J]. Front Plant Sci, 2023, 14: 1088285. |
| 6 | Wang B, Wang YK, Yuan X, et al. Comparative transcriptomic analysis provides key genetic resources in clove basil (Ocimum gratissimum) under cadmium stress [J]. Front Genet, 2023, 14: 1224140. |
| 7 | 郭向阳. 6种食用芳香植物挥发性成分的GC-MS/GC-O分析 [J]. 农业工程学报, 2019, 35(18): 299-307. |
| Guo XY. Analysis of volatile compositions in six edible fragrant plants by GC-MS/GC-O technology [J]. Trans Chin Soc Agric Eng, 2019, 35(18): 299-307. | |
| 8 | Hu ZY, Zhang YF, He Y, et al. Full-length transcriptome assembly of Italian ryegrass root integrated with RNA-seq to identify genes in response to plant cadmium stress [J]. Int J Mol Sci, 2020, 21(3): 1067. |
| 9 | Feng SJ, Shen YH, Xu HN, et al. RNA-seq identification of Cd responsive transporters provides insights into the association of oxidation resistance and Cd accumulation in Cucumis sativus L [J]. Antioxidants (Basel), 2021, 10(12): 1973. |
| 10 | He F, Liu QQ, Zheng L, et al. RNA-seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress [J]. Front Plant Sci, 2015, 6: 1136. |
| 11 | Li YK, Ding LH, Zhou M, et al. Transcriptional regulatory network of plant cadmium stress response [J]. Int J Mol Sci, 2023, 24(5): 4378. |
| 12 | 张晓娜, 朴春兰, 董友魁, 等. 大豆根系应答重金属Cd胁迫的转录组分析 [J]. 应用生态学报, 2017, 28(5): 1633-1641. |
| Zhang XN, Piao CL, Dong YK, et al. Transcriptome analysis of response to heavy metal Cd stress in soybean root [J]. Chin J Appl Ecol, 2017, 28(5): 1633-1641. | |
| 13 | Liu ZC, Zhou LZ, Gan CC, et al. Transcriptomic analysis reveals key genes and pathways corresponding to Cd and Pb in the hyperaccumulator Arabis paniculata [J]. Ecotoxicol Environ Saf, 2023, 254: 114757. |
| 14 | 肖艳辉, 李应文, 邹碧, 等. 钝化剂抑制南方污染农田籽粒苋吸收重金属的效应研究 [J]. 生态环境学报, 2021, 30(4): 825-833. |
| Xiao YH, Li YW, Zou B, et al. Reduction of heavy metal uptake by amaranth by 3 soil amendments in contaminated farmland of South China [J]. Ecol Environ Sci, 2021, 30(4): 825-833. | |
| 15 | 王斌, 袁晓, 蒋园园, 等. bHLH96的克隆及其在薄荷萜烯生物合成调控中的功能 [J]. 生物技术通报, 2024, 40(1): 281-293. |
| Wang B, Yuan X, Jiang YY, et al. Cloning of bHLH96 gene and its roles in regulating the biosynthesis of peppermint terpenes [J]. Biotechnol Bull, 2024, 40(1): 281-293. | |
| 16 | Geng YY, Qin LK, Liu YN, et al. Ultrastructure observation and transcriptome analysis of Chinese chestnut (Castanea mollissima Blume) seeds in response to water loss [J]. Food Biosci, 2021, 42: 101095. |
| 17 | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method [J]. Nat Protoc, 2008, 3(6): 1101-1108. |
| 18 | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis [J]. BMC Bioinformatics, 2008, 9: 559. |
| 19 | 宋琳, 刘梦宇, 周航, 等. 镉砷胁迫下玉米幼苗的生理响应和抗氧化系统特征 [J]. 农业环境科学学报, 2024, 43(4): 752-762. |
| Song L, Liu MY, Zhou H, et al. Physiological response and antioxidant system characteristics of maize (Zea mays L.) seedlings under cadmium and arsenic stress [J]. J Agro Environ Sci, 2024, 43(4): 752-762. | |
| 20 | Xu J, Yin HX, Liu XJ, et al. Salt affects plant Cd-stress responses by modulating growth and Cd accumulation [J]. Planta, 2010, 231(2): 449-459. |
| 21 | 张慧, 张欣雨, 袁旭, 等. 烟草叶片响应镉胁迫的差异表达基因鉴定及分析 [J]. 作物学报, 2024, 50(4): 944-956. |
| Zhang H, Zhang XY, Yuan X, et al. Transcriptome analysis of tobacco in response to cadmium stress [J]. Acta Agron Sin, 2024, 50(4): 944-956. | |
| 22 | Yan JY, Wu XZ, Li T, et al. Effect and mechanism of nano-materials on plant resistance to cadmium toxicity: a review [J]. Ecotoxicol Environ Saf, 2023, 266: 115576. |
| 23 | Khanna K, Kohli SK, Ohri P, et al. Agroecotoxicological aspect of Cd in soil-plant system: uptake, translocation and amelioration strategies [J]. Environ Sci Pollut Res Int, 2022, 29(21): 30908-30934. |
| 24 | Yu XF, Liu YJ, Yang L, et al. Low concentrations of methyl jasmonate promote plant growth and mitigate Cd toxicity in Cosmos bipinnatus [J]. BMC Plant Biol, 2024, 24(1): 807. |
| 25 | 李源恒, 赵春莉, 郭宏亮, 等. 镉胁迫对黄秋英生理及富集特性的影响 [J]. 山东农业科学, 2024, 56(1): 81-90. |
| Li YH, Zhao CL, Guo HL, et al. Effects of cadmium stress on physiological and enrichment characteristics of Cosmos sulphureus [J]. Shandong Agric Sci, 2024, 56(1): 81-90. | |
| 26 | Tong WY, Ahmad Rafiee AR, Leong CR, et al. Development of sodium alginate-pectin biodegradable active food packaging film containing cinnamic acid [J]. Chemosphere, 2023, 336: 139212. |
| 27 | Barroso JP, de Almeida AA F, do Nascimento JL, et al. The damage caused by Cd toxicity to photosynthesis, cellular ultrastructure, antioxidant metabolism, and gene expression in young cacao plants are mitigated by high Mn doses in soil [J]. Environ Sci Pollut Res Int, 2023, 30(54): 115646-115665. |
| 28 | Kou M, Xiong J, Li M, et al. Interactive effects of Cd and Pb on the photosynthesis efficiency and antioxidant defense system of Capsicum annuum L [J]. Bull Environ Contam Toxicol, 2022, 108(5): 917-925. |
| 29 | Wang SQ, Dai HP, Cui S, et al. The effects of salinity and pH variation on hyperaccumulator Bidens pilosa L. accumulating cadmium with dynamic and real-time uptake of Cd2+ influx around its root apexes [J]. Environ Sci Pollut Res Int, 2023, 30(14): 41435-41444. |
| 30 | 刘琛, 林义成, 郭彬, 等. 丛枝菌根真菌通过改变植物基因表达水平和微生物群落结构促进红叶石楠对镉的吸收 [J]. 生物工程学报, 2022, 38(1): 287-302. |
| Liu C, Lin YC, Guo B, et al. Arbuscular mycorrhizal fungi enhanced cadmium uptake in Photinia frase through altering root transcriptomes and root-associated microbial communities [J]. Chin J Biotechnol, 2022, 38(1): 287-302. | |
| 31 | Khan A, Jie Z, Kong XJ, et al. Pre treatment of melatonin rescues cotton seedlings from cadmium toxicity by regulating key physio-biochemical and molecular pathways [J]. J Hazard Mater, 2023, 445: 130530. |
| 32 | Li X, Xu B, Sahito ZA, et al. Transcriptome analysis reveals cadmium exposure enhanced the isoquinoline alkaloid biosynthesis and disease resistance in Coptis chinensis [J]. Ecotoxicol Environ Saf, 2024, 271: 115940. |
| 33 | Song L, Huang SSC, Wise A, et al. A transcription factor hierarchy defines an environmental stress response network [J]. Science, 2016, 354(6312): aag1550. |
| 34 | Zhang HW, Lu LL. Transcription factors involved in plant responses to cadmium-induced oxidative stress [J]. Front Plant Sci, 2024, 15: 1397289. |
| 35 | Khan I, Asaf S, Jan R, et al. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.) [J]. Front Plant Sci, 2023, 14: 1100895. |
| 36 | Yao YN, He ZQ, Li XM, et al. Genome-wide identification of bHLH gene family and its response to cadmium stress in Populus × canescens [J]. PeerJ, 2024, 12: e17410. |
| 37 | Du XY, Fang LH, Li JX, et al. The TabHLH094-TaMYC8 complex mediates the cadmium response in wheat [J]. Mol Breed, 2023, 43(7): 57. |
| [1] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [2] | 王斌, 袁晓, 蒋园园, 王玉昆, 肖艳辉, 何金明. bHLH96的克隆及其在薄荷萜烯生物合成调控中的功能[J]. 生物技术通报, 2024, 40(1): 281-293. |
| [3] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [4] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [5] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [6] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [7] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [8] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [9] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [10] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [11] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [12] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| [13] | 呼艳姣, 陈美凤, 强瑀, 李海燕, 刘静, 秦樊鑫. 镉胁迫下锌硒交互作用对水稻镉毒害的缓解机制[J]. 生物技术通报, 2022, 38(4): 143-152. |
| [14] | 祖国蔷, 胡哲, 王琪, 李光哲, 郝林. Burkholderia sp. GD17对水稻幼苗镉耐受的调节[J]. 生物技术通报, 2022, 38(4): 153-162. |
| [15] | 李兵娟, 郑璐, 沈仁芳, 兰平. 拟南芥RPP1A参与幼苗生长的蛋白质组学分析[J]. 生物技术通报, 2022, 38(2): 10-20. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||