








生物技术通报 ›› 2025, Vol. 41 ›› Issue (6): 256-268.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1105
• 研究报告 • 上一篇
瞿美玲1( ), 周思敏1, 张惊宇1, 何佳蔚1, 朱佳源1, 刘笑蓉1,2, 童巧珍1,2,3, 周日宝1,2,3,4(
), 周思敏1, 张惊宇1, 何佳蔚1, 朱佳源1, 刘笑蓉1,2, 童巧珍1,2,3, 周日宝1,2,3,4( ), 刘湘丹1,2,3,4(
), 刘湘丹1,2,3,4( )
)
收稿日期:2024-11-12
出版日期:2025-06-26
发布日期:2025-06-30
通讯作者:
:刘湘丹,博士,教授,博士生导师,研究方向 :中药资源与质量;E-mail: paeonia@hnucm.edu.cn作者简介:瞿美玲,女,硕士研究生,研究方向 :中药资源与质量;E-mail:2544018903@qq.com
基金资助:
QU Mei-ling1( ), ZHOU Si-min1, ZHANG Jing-yu1, HE Jia-wei1, ZHU Jia-yuan1, LIU Xiao-rong1,2, TONG Qiao-zhen1,2,3, ZHOU Ri-bao1,2,3,4(
), ZHOU Si-min1, ZHANG Jing-yu1, HE Jia-wei1, ZHU Jia-yuan1, LIU Xiao-rong1,2, TONG Qiao-zhen1,2,3, ZHOU Ri-bao1,2,3,4( ), LIU Xiang-dan1,2,3,4(
), LIU Xiang-dan1,2,3,4( )
)
Received:2024-11-12
Published:2025-06-26
Online:2025-06-30
摘要:
目的 研究灰毡毛忍冬bHLH转录因子家族成员的结构和功能,分析其表达特征,为后续深入研究灰毡毛忍冬bHLH基因家族在花发育过程中的功能奠定基础。 方法 基于灰毡毛忍冬两品种花转录组数据,筛选bHLH基因家族成员,利用在线工具进行生物信息学分析,获得灰毡毛忍冬两品种显著表达差异的基因,通过RT-qPCR验证bHLH的表达情况。 结果 共鉴定到55个bHLH转录因子家族成员,进化树分析显示灰毡毛忍冬bHLH蛋白与拟南芥有较高保守性,并基于进化树对部分bHLH蛋白的功能进行了预测。表达分析显示灰毡毛忍冬两品种bHLH基因存在显著差异,LmbHLH4/21/35在‘常规’中高表达,LmbHLH25在‘湘蕾’花期5-7中高表达。RT-qPCR结果显示,其相对表达量与转录组测序数据基本一致,证实了转录组测序结果的可靠性。 结论 推测LmbHLH4/21/15/25/35可能为参与调控灰毡毛忍冬花生长发育的关键转录因子,其为解析灰毡毛忍冬两品种花差异表型的分子机制提供了参考。
瞿美玲, 周思敏, 张惊宇, 何佳蔚, 朱佳源, 刘笑蓉, 童巧珍, 周日宝, 刘湘丹. 灰毡毛忍冬bHLH转录基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(6): 256-268.
QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides[J]. Biotechnology Bulletin, 2025, 41(6): 256-268.
引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| LmbHLH4 | ACGAATCGGTGGGTGTAAAG | CCCTTCTCCTCCTTTCAGATAAAT |
| LmbHLH14 | CTACTCTGCGCACCTTACTTC | GCACTCCTCCTCAGTTCTTTC |
| LmbHLH19 | GTAAGCCACTGGTGGATGTTAG | GGGTAAGAAGTTTGGAGGGATG |
| LmbHLH21 | CCGCCTTCAATAATCCCTACTC | CCGCTTGTTGTCTGTGTTTC |
| LmbHLH31 | TGTTGAGAGGACGCTTGATATG | GGAAGGATCCCAGGAGTAGTAA |
| LmbHLH34 | GTGGCAACTTCAGCAACAAA | CCCATTCCGTCACCGATAAA |
| LmbHLH35 | ACCTCAGCTGAAATGGAGAAG | TTCGTGGATGGGTAGCATAAC |
| LmbHLH39 | GCTTCTGTGGATCCCATGTT | GTTCCTGTGGGCTGGTTATT |
| LmbHLH55 | AGTCCATCCACCAGAGGTATTA | GAACAACGCCATGAGGAAGA |
| 18S | CTTCGGGATCGGAGTAATGA | GCGGAGTCCTAGAAGCAACA |
表1 RT-qPCR引物
Table 1 Primers in RT-qPCR
引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| LmbHLH4 | ACGAATCGGTGGGTGTAAAG | CCCTTCTCCTCCTTTCAGATAAAT |
| LmbHLH14 | CTACTCTGCGCACCTTACTTC | GCACTCCTCCTCAGTTCTTTC |
| LmbHLH19 | GTAAGCCACTGGTGGATGTTAG | GGGTAAGAAGTTTGGAGGGATG |
| LmbHLH21 | CCGCCTTCAATAATCCCTACTC | CCGCTTGTTGTCTGTGTTTC |
| LmbHLH31 | TGTTGAGAGGACGCTTGATATG | GGAAGGATCCCAGGAGTAGTAA |
| LmbHLH34 | GTGGCAACTTCAGCAACAAA | CCCATTCCGTCACCGATAAA |
| LmbHLH35 | ACCTCAGCTGAAATGGAGAAG | TTCGTGGATGGGTAGCATAAC |
| LmbHLH39 | GCTTCTGTGGATCCCATGTT | GTTCCTGTGGGCTGGTTATT |
| LmbHLH55 | AGTCCATCCACCAGAGGTATTA | GAACAACGCCATGAGGAAGA |
| 18S | CTTCGGGATCGGAGTAATGA | GCGGAGTCCTAGAAGCAACA |
基因名称 Gene name | 基因ID GeneID | 氨基酸数Number of amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定蛋白 Unstable protein | 亲水性 GRAVY | 脂肪指数 Fat index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| LmbHLH1 | TRINITY_DN43160_c3_g1 | 1 130 | 122 891.71 | 8.70 | 55.20 | -0.151 | 90.38 | 细胞核Nucleus |
| LmbHLH2 | TRINITY_DN45765_c0_g1 | 542 | 59 110.52 | 6.01 | 71.84 | -0.685 | 56.86 | 细胞核Nucleus |
| LmbHLH3 | TRINITY_DN44404_c4_g2 | 397 | 43 453.13 | 5.54 | 58.14 | -0.512 | 63.12 | 细胞核Nucleus |
| LmbHLH4 | TRINITY_DN45094_c5_g2 | 553 | 60 335.04 | 5.56 | 56.51 | -0.647 | 61.57 | 细胞核Nucleus |
| LmbHLH5 | TRINITY_DN47741_c4_g1 | 371 | 41 064.15 | 4.54 | 51.80 | -0.649 | 65.98 | 细胞核Nucleus |
| LmbHLH6 | TRINITY_DN46241_c4_g1 | 461 | 50 993.36 | 5.51 | 74.35 | -0.747 | 49.11 | 细胞核Nucleus |
| LmbHLH7 | TRINITY_DN39815_c0_g1 | 490 | 54 256.93 | 6.04 | 50.07 | -0.507 | 76.39 | 细胞核Nucleus |
| LmbHLH8 | TRINITY_DN42054_c1_g1 | 523 | 57 182.91 | 5.65 | 53.37 | -0.589 | 72.16 | 细胞核Nucleus |
| LmbHLH9 | TRINITY_DN42054_c1_g2 | 513 | 56 244.54 | 4.92 | 54.06 | -0.377 | 80.02 | 细胞核Nucleus |
| LmbHLH10 | TRINITY_DN32727_c0_g1 | 474 | 53 002.24 | 7.08 | 48.26 | -0.558 | 77.91 | 细胞核Nucleus |
| LmbHLH11 | TRINITY_DN40879_c0_g1 | 439 | 49 418.43 | 5.92 | 44.35 | -0.248 | 82.6 | 细胞核Nucleus |
| LmbHLH12 | TRINITY_DN42270_c3_g1 | 324 | 36 465.68 | 7.12 | 46.27 | -0.338 | 84.54 | 细胞核Nucleus |
| LmbHLH13 | TRINITY_DN46971_c4_g1 | 300 | 33 743.57 | 8.50 | 48.42 | -0.426 | 89.7 | 细胞核Nucleus |
| LmbHLH14 | TRINITY_DN42056_c4_g1 | 255 | 28 220.89 | 7.67 | 56.87 | -0.465 | 81.84 | 细胞核Nucleus |
| LmbHLH15 | TRINITY_DN44414_c1_g7 | 238 | 26 955.26 | 9.32 | 49.17 | -0.347 | 92.94 | 细胞核Nucleus |
| LmbHLH16 | TRINITY_DN44386_c1_g2 | 661 | 72 485.68 | 5.35 | 48.40 | -0.61 | 71.27 | 细胞核Nucleus |
| LmbHLH17 | TRINITY_DN36574_c0_g1 | 365 | 40 232.17 | 5.56 | 57.74 | -0.524 | 77.18 | 细胞核Nucleus |
| LmbHLH18 | TRINITY_DN42789_c3_g1 | 417 | 47 601.89 | 4.90 | 43.74 | -0.702 | 70.82 | 细胞核Nucleus |
| LmbHLH19 | TRINITY_DN20377_c0_g1 | 501 | 55 602.27 | 6.86 | 37.66 | -0.771 | 57.41 | 细胞核Nucleus |
| LmbHLH20 | TRINITY_DN46178_c3_g2 | 306 | 34 854.75 | 5.66 | 42.40 | -0.686 | 70.36 | 细胞核Nucleus |
| LmbHLH21 | TRINITY_DN42621_c1_g5 | 401 | 44 732.06 | 8.69 | 67.88 | -0.823 | 58.83 | 细胞核Nucleus |
| LmbHLH22 | TRINITY_DN18450_c0_g2 | 245 | 28 168.84 | 4.78 | 65.71 | -0.54 | 68.53 | 细胞核Nucleus |
| LmbHLH23 | TRINITY_DN40373_c2_g1 | 238 | 26 199.63 | 8.43 | 59.20 | -0.687 | 70.13 | 细胞核Nucleus |
| LmbHLH24 | TRINITY_DN43239_c3_g1 | 328 | 36 679.23 | 5.48 | 69.01 | -0.94 | 66.04 | 细胞核Nucleus |
| LmbHLH25 | TRINITY_DN42843_c1_g4 | 90 | 10 584.84 | 7.96 | 86.02 | -0.638 | 88.06 | 线粒体Mitochondria |
| LmbHLH26 | TRINITY_DN45117_c2_g1 | 182 | 20 540.08 | 9.69 | 47.10 | -0.267 | 106.54 | 细胞质Cytoplasm |
| LmbHLH27 | TRINITY_DN45385_c1_g1 | 146 | 16 139.3 | 8.97 | 50.05 | -0.446 | 74.18 | 细胞核Nucleus |
| LmbHLH28 | TRINITY_DN41621_c2_g4 | 136 | 15 036.15 | 9.35 | 45.97 | -0.511 | 82.43 | 细胞核Nucleus |
| LmbHLH29 | TRINITY_DN40447_c0_g2 | 365 | 40 187.54 | 9.01 | 64.24 | -0.938 | 55.89 | 细胞核Nucleus |
| LmbHLH30 | TRINITY_DN40447_c0_g3 | 493 | 54 438.76 | 6.91 | 47.43 | -0.758 | 66.92 | 细胞核Nucleus |
| LmbHLH31 | TRINITY_DN44673_c3_g1 | 519 | 56 623.36 | 6.00 | 53.66 | -0.64 | 62.95 | 细胞核Nucleus |
| LmbHLH32 | TRINITY_DN43098_c2_g1 | 449 | 46 998.16 | 6.45 | 48.96 | -0.458 | 67.19 | 细胞核Nucleus |
| LmbHLH33 | TRINITY_DN46720_c1_g5 | 386 | 42 523 | 6.08 | 42.60 | -0.451 | 76.01 | 细胞核Nucleus |
| LmbHLH34 | TRINITY_DN46720_c1_g2 | 398 | 43 975.37 | 5.69 | 47.20 | -0.668 | 66.18 | 细胞核Nucleus |
| LmbHLH35 | TRINITY_DN47048_c1_g1 | 329 | 36 614.72 | 6.14 | 43.61 | -0.75 | 70.24 | 细胞核Nucleus |
| LmbHLH36 | TRINITY_DN47048_c1_g5 | 361 | 39 136.52 | 8.20 | 53.55 | -0.658 | 62.99 | 细胞核Nucleus |
| LmbHLH37 | TRINITY_DN42352_c4_g1 | 378 | 42 296.4 | 4.78 | 53.80 | -0.839 | 64.29 | 细胞核Nucleus |
| LmbHLH38 | TRINITY_DN35329_c0_g1 | 282 | 31 833.52 | 5.60 | 47.70 | -0.649 | 60.5 | 细胞核Nucleus |
| LmbHLH39 | TRINITY_DN44560_c7_g3 | 244 | 27 872.51 | 8.37 | 46.42 | -0.946 | 62.75 | 细胞核Nucleus |
| LmbHLH40 | TRINITY_DN45264_c3_g1 | 114 | 13 132.32 | 10.45 | 54.28 | -0.275 | 83.77 | 叶绿体Chloroplasts |
| LmbHLH41 | TRINITY_DN46281_c1_g1 | 256 | 28 313.2 | 8.91 | 54.05 | -0.511 | 58.75 | 细胞核Nucleus |
| LmbHLH42 | TRINITY_DN2359_c0_g1 | 250 | 28 089.1 | 7.86 | 54.39 | -0.34 | 77.52 | 细胞核Nucleus |
| LmbHLH43 | TRINITY_DN44645_c1_g1 | 212 | 24 266.36 | 6.04 | 60.17 | -0.758 | 74.58 | 细胞核Nucleus |
| LmbHLH44 | TRINITY_DN44510_c4_g1 | 325 | 36 719.95 | 5.52 | 68.76 | -0.625 | 72 | 细胞核Nucleus |
| LmbHLH45 | TRINITY_DN22004_c0_g1 | 303 | 34 475.81 | 5.75 | 69.49 | -0.537 | 70.46 | 细胞核Nucleus |
| LmbHLH46 | TRINITY_DN42286_c0_g1 | 307 | 34 581.25 | 6.61 | 84.31 | -0.396 | 80.16 | 细胞核Nucleus |
| LmbHLH47 | TRINITY_DN38245_c0_g2 | 286 | 32 549.83 | 8.48 | 66.05 | -0.52 | 78.15 | 细胞核Nucleus |
| LmbHLH48 | TRINITY_DN36940_c0_g1 | 218 | 24 902.56 | 9.56 | 54.37 | -0.512 | 101.83 | 细胞质Cytoplasm |
| LmbHLH49 | TRINITY_DN44940_c4_g1 | 256 | 29 471.48 | 9.36 | 61.70 | -0.796 | 80.66 | 细胞核Nucleus |
| LmbHLH50 | TRINITY_DN36667_c0_g1 | 179 | 20 359.5 | 9.03 | 51.14 | -0.309 | 98.49 | 细胞核Nucleus |
| LmbHLH51 | TRINITY_DN44808_c6_g1 | 180 | 20 647.17 | 9.49 | 36.82 | -0.32 | 95.83 | 细胞核Nucleus |
| LmbHLH52 | TRINITY_DN45353_c0_g1 | 279 | 30 516.07 | 5.32 | 54.07 | -0.574 | 72.33 | 细胞核Nucleus |
| LmbHLH53 | TRINITY_DN47214_c0_g2 | 441 | 49 404.11 | 5.79 | 43.81 | -0.53 | 77.32 | 细胞核Nucleus |
| LmbHLH54 | TRINITY_DN47214_c0_g1 | 532 | 58 538.46 | 5.78 | 41.63 | -0.398 | 78.97 | 液泡膜Vacuole |
| LmbHLH55 | TRINITY_DN45078_c1_g2 | 884 | 96 521.93 | 6.24 | 38.58 | -0.355 | 81.26 | 细胞核Nucleus |
表2 灰毡毛忍冬bHLH基因家族的基本理化性质
Table 2 Basic physicochemical properties of the bHLH gene family of L. macranthoides
基因名称 Gene name | 基因ID GeneID | 氨基酸数Number of amino acids | 分子量 Molecular weight | 等电点 pI | 不稳定蛋白 Unstable protein | 亲水性 GRAVY | 脂肪指数 Fat index | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| LmbHLH1 | TRINITY_DN43160_c3_g1 | 1 130 | 122 891.71 | 8.70 | 55.20 | -0.151 | 90.38 | 细胞核Nucleus |
| LmbHLH2 | TRINITY_DN45765_c0_g1 | 542 | 59 110.52 | 6.01 | 71.84 | -0.685 | 56.86 | 细胞核Nucleus |
| LmbHLH3 | TRINITY_DN44404_c4_g2 | 397 | 43 453.13 | 5.54 | 58.14 | -0.512 | 63.12 | 细胞核Nucleus |
| LmbHLH4 | TRINITY_DN45094_c5_g2 | 553 | 60 335.04 | 5.56 | 56.51 | -0.647 | 61.57 | 细胞核Nucleus |
| LmbHLH5 | TRINITY_DN47741_c4_g1 | 371 | 41 064.15 | 4.54 | 51.80 | -0.649 | 65.98 | 细胞核Nucleus |
| LmbHLH6 | TRINITY_DN46241_c4_g1 | 461 | 50 993.36 | 5.51 | 74.35 | -0.747 | 49.11 | 细胞核Nucleus |
| LmbHLH7 | TRINITY_DN39815_c0_g1 | 490 | 54 256.93 | 6.04 | 50.07 | -0.507 | 76.39 | 细胞核Nucleus |
| LmbHLH8 | TRINITY_DN42054_c1_g1 | 523 | 57 182.91 | 5.65 | 53.37 | -0.589 | 72.16 | 细胞核Nucleus |
| LmbHLH9 | TRINITY_DN42054_c1_g2 | 513 | 56 244.54 | 4.92 | 54.06 | -0.377 | 80.02 | 细胞核Nucleus |
| LmbHLH10 | TRINITY_DN32727_c0_g1 | 474 | 53 002.24 | 7.08 | 48.26 | -0.558 | 77.91 | 细胞核Nucleus |
| LmbHLH11 | TRINITY_DN40879_c0_g1 | 439 | 49 418.43 | 5.92 | 44.35 | -0.248 | 82.6 | 细胞核Nucleus |
| LmbHLH12 | TRINITY_DN42270_c3_g1 | 324 | 36 465.68 | 7.12 | 46.27 | -0.338 | 84.54 | 细胞核Nucleus |
| LmbHLH13 | TRINITY_DN46971_c4_g1 | 300 | 33 743.57 | 8.50 | 48.42 | -0.426 | 89.7 | 细胞核Nucleus |
| LmbHLH14 | TRINITY_DN42056_c4_g1 | 255 | 28 220.89 | 7.67 | 56.87 | -0.465 | 81.84 | 细胞核Nucleus |
| LmbHLH15 | TRINITY_DN44414_c1_g7 | 238 | 26 955.26 | 9.32 | 49.17 | -0.347 | 92.94 | 细胞核Nucleus |
| LmbHLH16 | TRINITY_DN44386_c1_g2 | 661 | 72 485.68 | 5.35 | 48.40 | -0.61 | 71.27 | 细胞核Nucleus |
| LmbHLH17 | TRINITY_DN36574_c0_g1 | 365 | 40 232.17 | 5.56 | 57.74 | -0.524 | 77.18 | 细胞核Nucleus |
| LmbHLH18 | TRINITY_DN42789_c3_g1 | 417 | 47 601.89 | 4.90 | 43.74 | -0.702 | 70.82 | 细胞核Nucleus |
| LmbHLH19 | TRINITY_DN20377_c0_g1 | 501 | 55 602.27 | 6.86 | 37.66 | -0.771 | 57.41 | 细胞核Nucleus |
| LmbHLH20 | TRINITY_DN46178_c3_g2 | 306 | 34 854.75 | 5.66 | 42.40 | -0.686 | 70.36 | 细胞核Nucleus |
| LmbHLH21 | TRINITY_DN42621_c1_g5 | 401 | 44 732.06 | 8.69 | 67.88 | -0.823 | 58.83 | 细胞核Nucleus |
| LmbHLH22 | TRINITY_DN18450_c0_g2 | 245 | 28 168.84 | 4.78 | 65.71 | -0.54 | 68.53 | 细胞核Nucleus |
| LmbHLH23 | TRINITY_DN40373_c2_g1 | 238 | 26 199.63 | 8.43 | 59.20 | -0.687 | 70.13 | 细胞核Nucleus |
| LmbHLH24 | TRINITY_DN43239_c3_g1 | 328 | 36 679.23 | 5.48 | 69.01 | -0.94 | 66.04 | 细胞核Nucleus |
| LmbHLH25 | TRINITY_DN42843_c1_g4 | 90 | 10 584.84 | 7.96 | 86.02 | -0.638 | 88.06 | 线粒体Mitochondria |
| LmbHLH26 | TRINITY_DN45117_c2_g1 | 182 | 20 540.08 | 9.69 | 47.10 | -0.267 | 106.54 | 细胞质Cytoplasm |
| LmbHLH27 | TRINITY_DN45385_c1_g1 | 146 | 16 139.3 | 8.97 | 50.05 | -0.446 | 74.18 | 细胞核Nucleus |
| LmbHLH28 | TRINITY_DN41621_c2_g4 | 136 | 15 036.15 | 9.35 | 45.97 | -0.511 | 82.43 | 细胞核Nucleus |
| LmbHLH29 | TRINITY_DN40447_c0_g2 | 365 | 40 187.54 | 9.01 | 64.24 | -0.938 | 55.89 | 细胞核Nucleus |
| LmbHLH30 | TRINITY_DN40447_c0_g3 | 493 | 54 438.76 | 6.91 | 47.43 | -0.758 | 66.92 | 细胞核Nucleus |
| LmbHLH31 | TRINITY_DN44673_c3_g1 | 519 | 56 623.36 | 6.00 | 53.66 | -0.64 | 62.95 | 细胞核Nucleus |
| LmbHLH32 | TRINITY_DN43098_c2_g1 | 449 | 46 998.16 | 6.45 | 48.96 | -0.458 | 67.19 | 细胞核Nucleus |
| LmbHLH33 | TRINITY_DN46720_c1_g5 | 386 | 42 523 | 6.08 | 42.60 | -0.451 | 76.01 | 细胞核Nucleus |
| LmbHLH34 | TRINITY_DN46720_c1_g2 | 398 | 43 975.37 | 5.69 | 47.20 | -0.668 | 66.18 | 细胞核Nucleus |
| LmbHLH35 | TRINITY_DN47048_c1_g1 | 329 | 36 614.72 | 6.14 | 43.61 | -0.75 | 70.24 | 细胞核Nucleus |
| LmbHLH36 | TRINITY_DN47048_c1_g5 | 361 | 39 136.52 | 8.20 | 53.55 | -0.658 | 62.99 | 细胞核Nucleus |
| LmbHLH37 | TRINITY_DN42352_c4_g1 | 378 | 42 296.4 | 4.78 | 53.80 | -0.839 | 64.29 | 细胞核Nucleus |
| LmbHLH38 | TRINITY_DN35329_c0_g1 | 282 | 31 833.52 | 5.60 | 47.70 | -0.649 | 60.5 | 细胞核Nucleus |
| LmbHLH39 | TRINITY_DN44560_c7_g3 | 244 | 27 872.51 | 8.37 | 46.42 | -0.946 | 62.75 | 细胞核Nucleus |
| LmbHLH40 | TRINITY_DN45264_c3_g1 | 114 | 13 132.32 | 10.45 | 54.28 | -0.275 | 83.77 | 叶绿体Chloroplasts |
| LmbHLH41 | TRINITY_DN46281_c1_g1 | 256 | 28 313.2 | 8.91 | 54.05 | -0.511 | 58.75 | 细胞核Nucleus |
| LmbHLH42 | TRINITY_DN2359_c0_g1 | 250 | 28 089.1 | 7.86 | 54.39 | -0.34 | 77.52 | 细胞核Nucleus |
| LmbHLH43 | TRINITY_DN44645_c1_g1 | 212 | 24 266.36 | 6.04 | 60.17 | -0.758 | 74.58 | 细胞核Nucleus |
| LmbHLH44 | TRINITY_DN44510_c4_g1 | 325 | 36 719.95 | 5.52 | 68.76 | -0.625 | 72 | 细胞核Nucleus |
| LmbHLH45 | TRINITY_DN22004_c0_g1 | 303 | 34 475.81 | 5.75 | 69.49 | -0.537 | 70.46 | 细胞核Nucleus |
| LmbHLH46 | TRINITY_DN42286_c0_g1 | 307 | 34 581.25 | 6.61 | 84.31 | -0.396 | 80.16 | 细胞核Nucleus |
| LmbHLH47 | TRINITY_DN38245_c0_g2 | 286 | 32 549.83 | 8.48 | 66.05 | -0.52 | 78.15 | 细胞核Nucleus |
| LmbHLH48 | TRINITY_DN36940_c0_g1 | 218 | 24 902.56 | 9.56 | 54.37 | -0.512 | 101.83 | 细胞质Cytoplasm |
| LmbHLH49 | TRINITY_DN44940_c4_g1 | 256 | 29 471.48 | 9.36 | 61.70 | -0.796 | 80.66 | 细胞核Nucleus |
| LmbHLH50 | TRINITY_DN36667_c0_g1 | 179 | 20 359.5 | 9.03 | 51.14 | -0.309 | 98.49 | 细胞核Nucleus |
| LmbHLH51 | TRINITY_DN44808_c6_g1 | 180 | 20 647.17 | 9.49 | 36.82 | -0.32 | 95.83 | 细胞核Nucleus |
| LmbHLH52 | TRINITY_DN45353_c0_g1 | 279 | 30 516.07 | 5.32 | 54.07 | -0.574 | 72.33 | 细胞核Nucleus |
| LmbHLH53 | TRINITY_DN47214_c0_g2 | 441 | 49 404.11 | 5.79 | 43.81 | -0.53 | 77.32 | 细胞核Nucleus |
| LmbHLH54 | TRINITY_DN47214_c0_g1 | 532 | 58 538.46 | 5.78 | 41.63 | -0.398 | 78.97 | 液泡膜Vacuole |
| LmbHLH55 | TRINITY_DN45078_c1_g2 | 884 | 96 521.93 | 6.24 | 38.58 | -0.355 | 81.26 | 细胞核Nucleus |
蛋白 Protein | α-螺旋α-helix (%) | β-转角 β-sheet (%) | 折叠延伸链 Turn loop (%) | 无规则卷曲 Random coil (%) | 蛋白 Protein | α-螺旋α-helix (%) | β-转角 β-sheet (%) | 折叠延伸链 Turn loop (%) | 无规则卷曲 Random coil (%) |
|---|---|---|---|---|---|---|---|---|---|
| LmbHLH1 | 46.02 | 0.00 | 1.86 | 52.12 | LmbHLH29 | 16.99 | 0.00 | 2.47 | 80.55 |
| LmbHLH2 | 13.84 | 0.00 | 0.74 | 85.42 | LmbHLH30 | 20.69 | 0.00 | 1.42 | 77.89 |
| LmbHLH3 | 18.14 | 0.00 | 0.76 | 81.11 | LmbHLH31 | 13.68 | 0.00 | 2.12 | 84.20 |
| LmbHLH4 | 14.29 | 0.00 | 0.72 | 84.99 | LmbHLH32 | 23.39 | 0.00 | 1.56 | 75.06 |
| LmbHLH5 | 19.95 | 0.00 | 2.96 | 77.09 | LmbHLH33 | 23.83 | 0.00 | 1.30 | 74.87 |
| LmbHLH6 | 14.75 | 0.00 | 3.04 | 82.21 | LmbHLH34 | 21.86 | 0.00 | 1.26 | 76.88 |
| LmbHLH7 | 30.41 | 0.00 | 8.98 | 60.61 | LmbHLH35 | 25.53 | 0.00 | 3.04 | 71.43 |
| LmbHLH8 | 22.56 | 0.00 | 5.93 | 71.51 | LmbHLH36 | 22.71 | 0.00 | 3.05 | 74.24 |
| LmbHLH9 | 17.54 | 0.00 | 7.02 | 75.44 | LmbHLH37 | 17.20 | 0.00 | 2.12 | 80.69 |
| LmbHLH10 | 30.38 | 0.00 | 8.44 | 61.18 | LmbHLH38 | 30.85 | 0.00 | 1.77 | 67.38 |
| LmbHLH11 | 30.30 | 0.00 | 10.93 | 58.77 | LmbHLH39 | 29.10 | 0.00 | 2.05 | 68.85 |
| LmbHLH12 | 35.80 | 0.00 | 5.25 | 58.95 | LmbHLH40 | 39.47 | 0.00 | 9.65 | 50.88 |
| LmbHLH13 | 44.67 | 0.00 | 6.67 | 48.67 | LmbHLH41 | 28.91 | 0.00 | 1.17 | 69.92 |
| LmbHLH14 | 38.43 | 0.00 | 9.41 | 52.16 | LmbHLH42 | 31.20 | 0.00 | 0.00 | 68.80 |
| LmbHLH15 | 44.12 | 0.00 | 7.98 | 47.90 | LmbHLH43 | 31.13 | 0.00 | 0.47 | 68.40 |
| LmbHLH16 | 27.23 | 0.00 | 8.17 | 64.60 | LmbHLH44 | 32.62 | 0.00 | 6.46 | 60.92 |
| LmbHLH17 | 29.86 | 0.00 | 5.21 | 64.93 | LmbHLH45 | 34.98 | 0.00 | 7.92 | 57.10 |
| LmbHLH18 | 32.13 | 0.00 | 6.24 | 61.63 | LmbHLH46 | 35.18 | 0.00 | 6.51 | 58.31 |
| LmbHLH19 | 13.97 | 0.00 | 4.19 | 81.84 | LmbHLH47 | 34.27 | 0.00 | 7.34 | 58.39 |
| LmbHLH20 | 28.76 | 0.00 | 1.96 | 69.28 | LmbHLH48 | 41.28 | 0.00 | 10.55 | 48.17 |
| LmbHLH21 | 21.95 | 0.00 | 0.75 | 77.31 | LmbHLH49 | 35.94 | 0.00 | 8.98 | 55.08 |
| LmbHLH22 | 32.65 | 0.00 | 0.82 | 66.53 | LmbHLH50 | 50.28 | 0.00 | 11.17 | 38.55 |
| LmbHLH23 | 40.34 | 0.00 | 0.00 | 59.66 | LmbHLH51 | 49.44 | 0.00 | 11.67 | 38.89 |
| LmbHLH24 | 25.00 | 0.00 | 2.44 | 72.56 | LmbHLH52 | 22.94 | 0.00 | 5.73 | 71.33 |
| LmbHLH25 | 62.37 | 0.00 | 1.08 | 36.56 | LmbHLH53 | 32.65 | 0.00 | 3.63 | 63.72 |
| LmbHLH26 | 48.35 | 0.00 | 9.34 | 42.31 | LmbHLH54 | 29.14 | 0.00 | 4.89 | 65.98 |
| LmbHLH27 | 29.45 | 0.00 | 8.22 | 62.33 | LmbHLH55 | 22.85 | 0.00 | 5.88 | 71.27 |
| LmbHLH28 | 31.62 | 0.00 | 9.56 | 58.82 |
表3 LmbHLH蛋白二级结构分析
Table 3 Secondary structure analysis of LmbHLH protein
蛋白 Protein | α-螺旋α-helix (%) | β-转角 β-sheet (%) | 折叠延伸链 Turn loop (%) | 无规则卷曲 Random coil (%) | 蛋白 Protein | α-螺旋α-helix (%) | β-转角 β-sheet (%) | 折叠延伸链 Turn loop (%) | 无规则卷曲 Random coil (%) |
|---|---|---|---|---|---|---|---|---|---|
| LmbHLH1 | 46.02 | 0.00 | 1.86 | 52.12 | LmbHLH29 | 16.99 | 0.00 | 2.47 | 80.55 |
| LmbHLH2 | 13.84 | 0.00 | 0.74 | 85.42 | LmbHLH30 | 20.69 | 0.00 | 1.42 | 77.89 |
| LmbHLH3 | 18.14 | 0.00 | 0.76 | 81.11 | LmbHLH31 | 13.68 | 0.00 | 2.12 | 84.20 |
| LmbHLH4 | 14.29 | 0.00 | 0.72 | 84.99 | LmbHLH32 | 23.39 | 0.00 | 1.56 | 75.06 |
| LmbHLH5 | 19.95 | 0.00 | 2.96 | 77.09 | LmbHLH33 | 23.83 | 0.00 | 1.30 | 74.87 |
| LmbHLH6 | 14.75 | 0.00 | 3.04 | 82.21 | LmbHLH34 | 21.86 | 0.00 | 1.26 | 76.88 |
| LmbHLH7 | 30.41 | 0.00 | 8.98 | 60.61 | LmbHLH35 | 25.53 | 0.00 | 3.04 | 71.43 |
| LmbHLH8 | 22.56 | 0.00 | 5.93 | 71.51 | LmbHLH36 | 22.71 | 0.00 | 3.05 | 74.24 |
| LmbHLH9 | 17.54 | 0.00 | 7.02 | 75.44 | LmbHLH37 | 17.20 | 0.00 | 2.12 | 80.69 |
| LmbHLH10 | 30.38 | 0.00 | 8.44 | 61.18 | LmbHLH38 | 30.85 | 0.00 | 1.77 | 67.38 |
| LmbHLH11 | 30.30 | 0.00 | 10.93 | 58.77 | LmbHLH39 | 29.10 | 0.00 | 2.05 | 68.85 |
| LmbHLH12 | 35.80 | 0.00 | 5.25 | 58.95 | LmbHLH40 | 39.47 | 0.00 | 9.65 | 50.88 |
| LmbHLH13 | 44.67 | 0.00 | 6.67 | 48.67 | LmbHLH41 | 28.91 | 0.00 | 1.17 | 69.92 |
| LmbHLH14 | 38.43 | 0.00 | 9.41 | 52.16 | LmbHLH42 | 31.20 | 0.00 | 0.00 | 68.80 |
| LmbHLH15 | 44.12 | 0.00 | 7.98 | 47.90 | LmbHLH43 | 31.13 | 0.00 | 0.47 | 68.40 |
| LmbHLH16 | 27.23 | 0.00 | 8.17 | 64.60 | LmbHLH44 | 32.62 | 0.00 | 6.46 | 60.92 |
| LmbHLH17 | 29.86 | 0.00 | 5.21 | 64.93 | LmbHLH45 | 34.98 | 0.00 | 7.92 | 57.10 |
| LmbHLH18 | 32.13 | 0.00 | 6.24 | 61.63 | LmbHLH46 | 35.18 | 0.00 | 6.51 | 58.31 |
| LmbHLH19 | 13.97 | 0.00 | 4.19 | 81.84 | LmbHLH47 | 34.27 | 0.00 | 7.34 | 58.39 |
| LmbHLH20 | 28.76 | 0.00 | 1.96 | 69.28 | LmbHLH48 | 41.28 | 0.00 | 10.55 | 48.17 |
| LmbHLH21 | 21.95 | 0.00 | 0.75 | 77.31 | LmbHLH49 | 35.94 | 0.00 | 8.98 | 55.08 |
| LmbHLH22 | 32.65 | 0.00 | 0.82 | 66.53 | LmbHLH50 | 50.28 | 0.00 | 11.17 | 38.55 |
| LmbHLH23 | 40.34 | 0.00 | 0.00 | 59.66 | LmbHLH51 | 49.44 | 0.00 | 11.67 | 38.89 |
| LmbHLH24 | 25.00 | 0.00 | 2.44 | 72.56 | LmbHLH52 | 22.94 | 0.00 | 5.73 | 71.33 |
| LmbHLH25 | 62.37 | 0.00 | 1.08 | 36.56 | LmbHLH53 | 32.65 | 0.00 | 3.63 | 63.72 |
| LmbHLH26 | 48.35 | 0.00 | 9.34 | 42.31 | LmbHLH54 | 29.14 | 0.00 | 4.89 | 65.98 |
| LmbHLH27 | 29.45 | 0.00 | 8.22 | 62.33 | LmbHLH55 | 22.85 | 0.00 | 5.88 | 71.27 |
| LmbHLH28 | 31.62 | 0.00 | 9.56 | 58.82 |
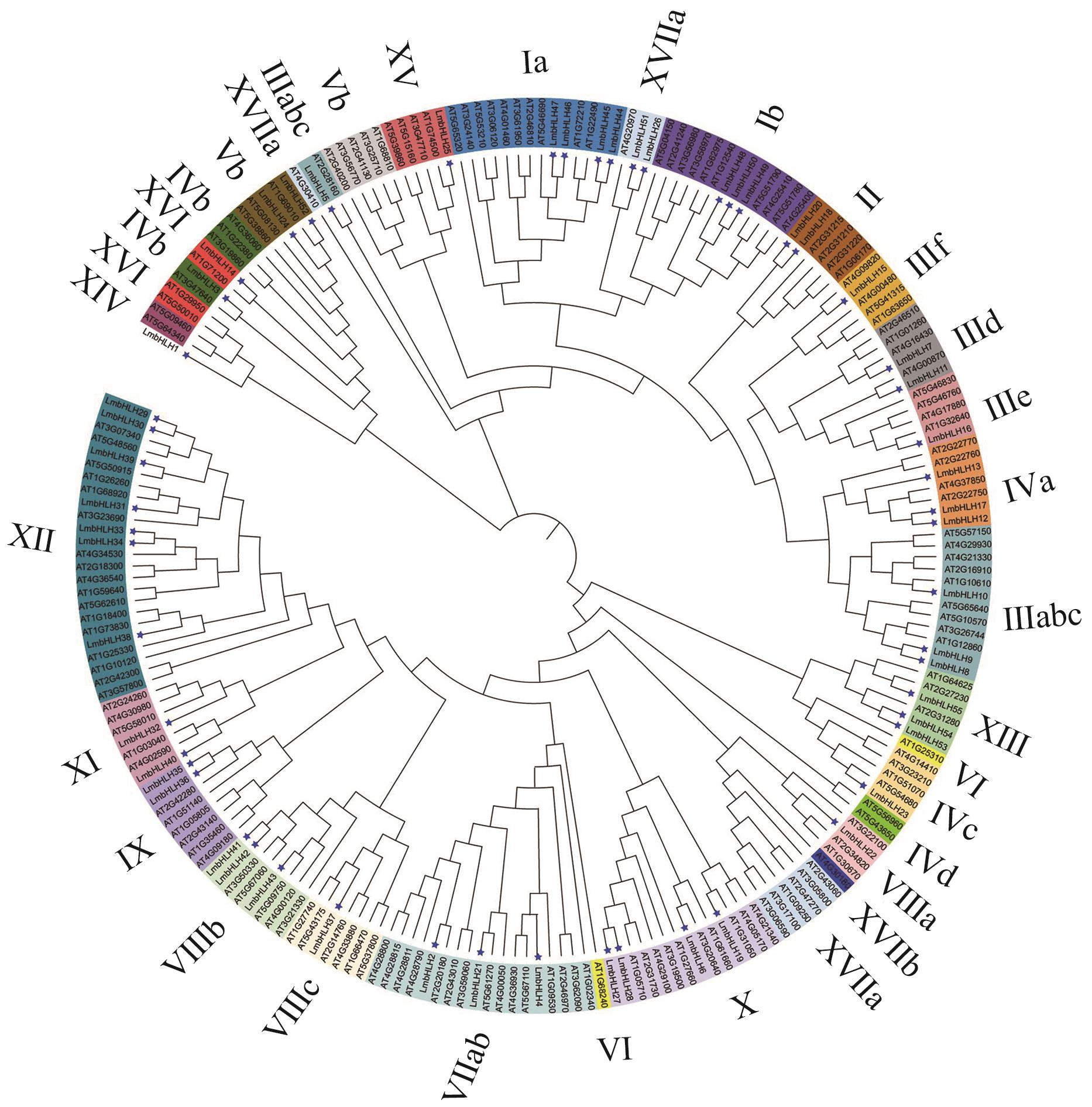
图5 灰毡毛忍冬、拟南芥bHLH转录因子家族系统进化树蓝色五角星表示灰毡毛忍冬bHLH转录因子家族成员;其余皆为拟南芥bHLH转录因子家族成员
Fig. 5 Phylogenetic tree of L. macranthoides and A. thalianabHLH transcription factor familyBlue pentagram indicates a member of the bHLH transcription factor family of L. macranthoides; the rest are members of the ArabidopsisbHLH transcription factor family
基因名称 Gene name | 基因ID Gene ID | 功能 Function | 组别 Peer group | 参考文献Reference |
|---|---|---|---|---|
| LmbHLH44 | At4G01460 | 参与种子休眠过程 | Ia | [ |
| LmbHLH45/46/47 | At3G24140 | 促进气孔保护细胞的分化 | Ia | [ |
| LmbHLH16 | At1G32640 | 参与茉莉酸、脱落酸和光信号通路;调节黄酮类化合物和色氨酸的生物合成 | IIIe | [ |
| LmbHLH15 | At1G63650 | 调节花青素生物合成、毛状体和根毛发育 | IIIf | [ |
| LmbHLH7/11 | At4G16430 | 抑制MYC2激活的叶片衰老,负调控JA应答 | IIId | [ |
| LmbHLH4 | At2G43010 | 调控开花时间 | VIIa | [ |
| LmbHLH21 | At4G36930 | 响应雌蕊的生长素生物合成 | VIIb | [ |
| LmbHLH25 | At3G47710 | 调控花冠形态,促进叶绿素积累 | XⅤ | [ |
| LmbHLH23 | At4G14410 | 调节缺铁反应 | IVc | [ |
| LmbHLH6 | At1G61660 | 调节多重反应,提高应激耐受性 | X | [ |
| LmbHLH38 | At1G18400 | 光周期开花的正调节剂 | XII | [ |
| LmbHLH37 | At4G33880 | 参与根毛发育 | VIIIc | [ |
| LmbHLH35/36 | At1G51140 | 抑制(ABA)脱落酸分解代谢 | IX | [ |
| LmbHLH35/36/26/51 | At1G35460 | 控制光周期开花调节因子CO的表达 | IX | [ |
表4 以同源AtbHLH基因为参考的LmbHLH基因功能推测
Table 4 Functional prediction of LmbHLH gene based on homologous AtbHLH gene
基因名称 Gene name | 基因ID Gene ID | 功能 Function | 组别 Peer group | 参考文献Reference |
|---|---|---|---|---|
| LmbHLH44 | At4G01460 | 参与种子休眠过程 | Ia | [ |
| LmbHLH45/46/47 | At3G24140 | 促进气孔保护细胞的分化 | Ia | [ |
| LmbHLH16 | At1G32640 | 参与茉莉酸、脱落酸和光信号通路;调节黄酮类化合物和色氨酸的生物合成 | IIIe | [ |
| LmbHLH15 | At1G63650 | 调节花青素生物合成、毛状体和根毛发育 | IIIf | [ |
| LmbHLH7/11 | At4G16430 | 抑制MYC2激活的叶片衰老,负调控JA应答 | IIId | [ |
| LmbHLH4 | At2G43010 | 调控开花时间 | VIIa | [ |
| LmbHLH21 | At4G36930 | 响应雌蕊的生长素生物合成 | VIIb | [ |
| LmbHLH25 | At3G47710 | 调控花冠形态,促进叶绿素积累 | XⅤ | [ |
| LmbHLH23 | At4G14410 | 调节缺铁反应 | IVc | [ |
| LmbHLH6 | At1G61660 | 调节多重反应,提高应激耐受性 | X | [ |
| LmbHLH38 | At1G18400 | 光周期开花的正调节剂 | XII | [ |
| LmbHLH37 | At4G33880 | 参与根毛发育 | VIIIc | [ |
| LmbHLH35/36 | At1G51140 | 抑制(ABA)脱落酸分解代谢 | IX | [ |
| LmbHLH35/36/26/51 | At1G35460 | 控制光周期开花调节因子CO的表达 | IX | [ |

图6 不同花期LmbHLH基因表达量热图A:花期1;B:花期2;C:花期3;D:花期4;E:花期5;F:花期6;G:花期7。Lm_xl:‘湘蕾’灰毡毛忍冬;Lm_wt:‘常规’灰毡毛忍冬。下同
Fig. 6 Heat map of LmbHLH gene expressions at different flowering stagesA: Flowering period 1. B: Flowering period 2. C: Flowering period 3. D: Flowering period 4. E: Flowering period 5. F: Flowering period 6. G: Flowering period 7. Lm_xl.: 'Xianglei' L. macranthoides; Lm_wt: 'Changgui' L. macranthoides. The same below
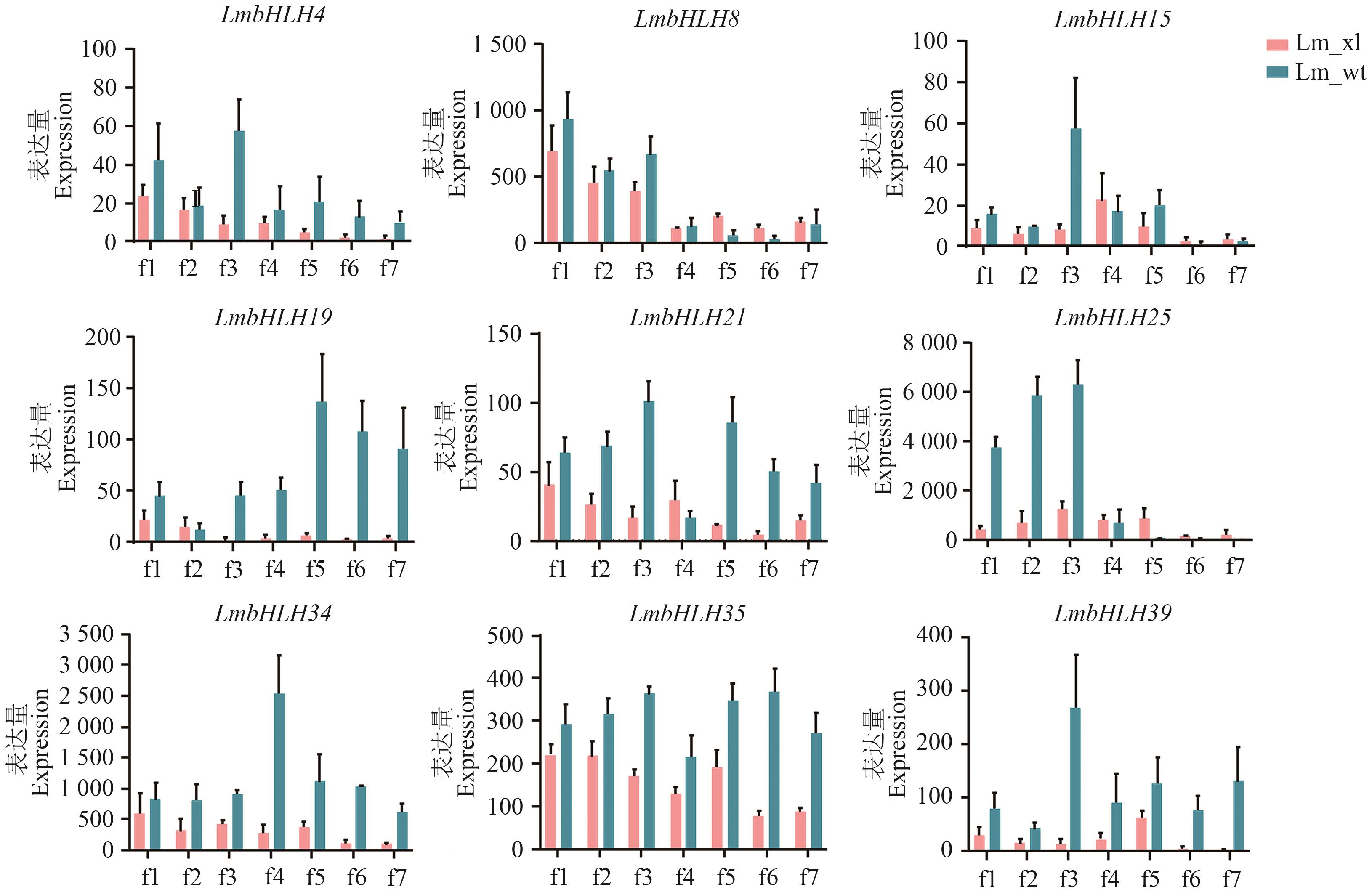
图7 灰毡毛忍冬两品种间不同花期bHLH表达量f1-f7:花期1-7
Fig. 7 Expressions of bHLH in different flowering stages between the two varieties of L. macranthoidesf1-f7: Flowering period 1-7
| 1 | 中国科学院中国植物志编辑委员会,丘华兴,等. 中国植物志[M]. 北京: 科学出版社, 1988. |
| Chinese Flora Editing Committee, Chinese Academy of Sciences,Qiu HX, et al. Flora of China [M]. Beijing: Science Press, 1988. | |
| 2 | 国家药典委员会. 中华人民共和国药典-三部: 2020年版 [M]. 北京: 中国医药科技出版社, 2020. |
| National Pharmacopoeia Commission. People’s republic of China (PRC) pharmacopoeia-part III: 2020 edition [M]. Beijing: China Medical Science Press, 2020. | |
| 3 | 龙丽君, 曾慧杰, 乔中全, 等. 灰毡毛忍冬AGL12基因克隆及互作蛋白鉴定 [J]. 药学学报, 2024, 59(5): 1458-1466. |
| Long LJ, Zeng HJ, Qiao ZQ, et al. Cloning and interacted protein identification of AGL12 gene from Lonicera macranthoides [J]. Acta Pharm Sin, 2024, 59(5): 1458-1466. | |
| 4 | 陈仕高, 刘朝敏, 杨美森. 山银花幼龄期快速丰产技术 [J]. 基层农技推广, 2023, 11(11): 96-99. |
| Chen SG, Liu CM, Yang MS. Rapid high-yield techniques of Lonicera japonica in young age [J]. Prim Agric Technol Ext, 2023, 11(11): 96-99. | |
| 5 | 易刚强, 蔡嘉洛, 朱贻霖, 等. 灰毡毛忍冬MADS-box基因家族AGL15基因的克隆、生物信息学和表达分析 [J]. 中草药, 2016, 47(4): 640-647. |
| Yi GQ, Cai JL, Zhu YL, et al. Cloning of MADS-box gene AGL15 from Lonicera macranthoides and analysis on its bioinformatics and expression [J]. Chin Tradit Herb Drugs, 2016, 47(4): 640-647. | |
| 6 | Xia Y, Chen WW, Xiang WB, et al. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in Lonicera japonica flower petals during colour-transition [J]. BMC Plant Biol, 2021, 21(1): 98. |
| 7 | Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling [J]. Plant Cell, 2003, 15(1): 63-78. |
| 8 | Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors [J]. Plant J, 2011, 66(1): 94-116. |
| 9 | Qian YC, Zhang TY, Yu Y, et al. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses [J]. Front Plant Sci, 2021, 12: 677611. |
| 10 | Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins [J]. J Mol Evol, 2003, 56(6): 742-750. |
| 11 | Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis [J]. Genome Res, 2001, 11(5): 754-770. |
| 12 | Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis [J]. Int J Mol Sci, 2021, 22(13): 7152. |
| 13 | Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins [J]. Cell, 1989, 56(5): 777-783. |
| 14 | Zhang F, Gonzalez A, Zhao MZ, et al. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis [J]. Development, 2003, 130(20): 4859-4869. |
| 15 | Nesi N, Jond C, Debeaujon I, et al. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed [J]. Plant Cell, 2001, 13(9): 2099-2114. |
| 16 | Nesi N, Debeaujon I, Jond C, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques [J]. Plant Cell, 2000, 12(10): 1863-1878. |
| 17 | Wang F, Gao YS, Liu YW, et al. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis [J]. New Phytol, 2019, 223(3): 1407-1419. |
| 18 | Ito S, Song YH, Josephson-Day AR, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis [J]. Proc Natl Acad Sci USA, 2012, 109(9): 3582-3587. |
| 19 | Waseem M, Li N, Su DD, et al. Overexpression of a basic helix-loop-helix transcription factor gene, SlbHLH22, promotes early flowering and accelerates fruit ripening in tomato (Solanum lycopersicum L.) [J]. Planta, 2019, 250(1): 173-185. |
| 20 | Chen Q, Li J, Yang FJ. Genome-wide analysis of the mads-box transcription factor family in Solanum melongena [J]. Int J Mol Sci, 2023, 24(1): 826. |
| 21 | Song MY, Wang HM, Wang Z, et al. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in fig (Ficus carica L.) [J]. Front Plant Sci, 2021, 12: 730692. |
| 22 | 刘畅宇, 陈勋, 陈娅, 等. 不同品种灰毡毛忍冬ACS3基因克隆、表达及生物信息学分析 [J]. 中草药, 2019, 50(9): 2154-2164. |
| Liu CY, Chen X, Chen Y, et al. Cloning, space-time expression and bioinformatics analysis of ACS3 gene from different species of Lonicera macranthoides [J]. Chin Tradit Herb Drugs, 2019, 50(9): 2154-2164. | |
| 23 | Hall BG. Building phylogenetic trees from molecular data with MEGA [J]. Mol Biol Evol, 2013, 30(5): 1229-1235. |
| 24 | Zhang Y, Gao WL, Li HT, et al. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.) [J]. BMC Genomics, 2020, 21(1): 483. |
| 25 | Shen TJ, Wen XP, Wen Z, et al. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.) [J]. Sci Hortic, 2021, 279: 109905. |
| 26 | Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants [J]. Mol Biol Evol, 2010, 27(4): 862-874. |
| 27 | Gao F, Dubos C. The Arabidopsis bHLH transcription factor family [J]. Trends Plant Sci, 2024, 29(6): 668-680. |
| 28 | Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity [J]. Mol Biol Evol, 2003, 20(5): 735-747. |
| 29 | Lefebvre V, North H, Frey A, et al. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy [J]. Plant J, 2006, 45(3): 309-319. |
| 30 | Qi TC, Huang H, Song SS, et al. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis [J]. Plant Cell, 2015, 27(6): 1620-1633. |
| 31 | Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity [J]. Trends Plant Sci, 2005, 10(2): 63-70. |
| 32 | Vinod Kumar S, Lucyshyn D, Jaeger KE, et al. Transcription factor PIF4 controls the thermosensory activation of flowering [J]. Nature, 2012, 484(7393): 242-245. |
| 33 | Groszmann M, Bylstra Y, Lampugnani ER, et al. Regulation of tissue-specific expression of SPATULA a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis [J]. J Exp Bot, 2010, 61(5): 1495-1508. |
| 34 | Mara CD, Huang TB, Irish VF. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling [J]. Plant Cell, 2010, 22(3): 690-702. |
| 35 | Zhang J, Liu B, Li MS, et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis [J]. Plant Cell, 2015, 27(3): 787-805. |
| 36 | Wang WS, Zhu J, Lu YT. Overexpression of AtbHLH112 suppresses lateral root emergence in Arabidopsis [J]. Funct Plant Biol, 2014, 41(4): 342-352. |
| 37 | Menand B, Yi KK, Jouannic S, et al. An ancient mechanism controls the development of cells with a rooting function in land plants [J]. Science, 2007, 316(5830): 1477-1480. |
| 38 | Liu WW, Tai HH, Li SS, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism [J]. New Phytol, 2014, 201(4): 1192-1204. |
| 39 | Zuo ZF, Lee HY, Kang HG. Basic helix-loop-helix transcription factors: regulators for plant growth development and abiotic stress responses [J]. Int J Mol Sci, 2023, 24(2): 1419. |
| 40 | Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis [J]. Plant Physiol, 2006, 141(4): 1167-1184. |
| 41 | Li JL, Wang T, Han J, et al. Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber [J]. BMC Plant Biol, 2020, 20(1): 272. |
| 42 | Khan I, Asaf S, Jan R, et al. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.) [J]. Front Plant Sci, 2023, 14: 1100895. |
| 43 | Zhou X, Liao YL, Kim SU, et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba [J]. Sci Rep, 2020, 10(1): 13723. |
| 44 | Zhang CH, Feng RC, Ma RJ, et al. Genome-wide analysis of basic helix-loop-helix superfamily members in peach [J]. PLoS One, 2018, 13(4): e0195974. |
| 45 | Liu JY, Chen NN, Chen F, et al. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera) [J]. BMC Genomics, 2014, 15: 281. |
| 46 | Liu SY, Zhang CB, Guo F, et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato [J]. BMC Plant Biol, 2022, 22(1): 616. |
| 47 | Gonzalez A, Zhao MZ, Leavitt JM, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings [J]. Plant J, 2008, 53(5): 814-827. |
| 48 | Yin J, Chang XX, Kasuga T, et al. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in Petunia [J]. Hortic Res, 2015, 2: 15059. |
| [1] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [2] | 罗嗣芳, 张祖铭, 谢丽芳, 郭紫晶, 陈兆星, 杨月华, 严翔, 张洪铭. 山金柑GATA基因家族全基因组鉴定及在果实发育中的表达分析[J]. 生物技术通报, 2025, 41(5): 218-230. |
| [3] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [4] | 赵婧, 郭茜, 李睿琦, 雷滢炀, 岳爱琴, 赵晋忠, 殷丛丛, 杜维俊, 牛景萍. 大豆GmGST基因簇基因序列分析及诱导表达分析[J]. 生物技术通报, 2025, 41(5): 129-140. |
| [5] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [6] | 刘鑫, 王嘉雯, 李进伟, 牟策, 杨盼盼, 明军, 徐雷锋. 兰州百合三个LdBBXs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(5): 186-196. |
| [7] | 孙天国, 衣兰, 秦旭洋, 乔梦雪, 谷新颖, 韩艺, 沙伟, 张梅娟, 马天意. 大白菜DABB基因家族的全基因组鉴定及盐碱胁迫下的表达分析[J]. 生物技术通报, 2025, 41(4): 156-165. |
| [8] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [9] | 王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187. |
| [10] | 黄金恒, 黄茜, 张家燕, 周新裕, 廖沛然, 杨全. 广金钱草C3H基因家族鉴定及不同品种表达分析[J]. 生物技术通报, 2025, 41(4): 243-255. |
| [11] | 班秋艳, 赵鑫月, 迟文静, 黎俊生, 王琼, 夏瑶, 梁丽云, 贺巍, 李叶云, 赵广山. 茶树光敏色素互作因子CsPIF3a的克隆及其与光温逆境的响应[J]. 生物技术通报, 2025, 41(4): 256-265. |
| [12] | 卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277. |
| [13] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [14] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [15] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||