








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 266-277.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0894
• 研究报告 • 上一篇
卢勇杰1( ), 夏海乾2, 李永铃1, 张文建3, 余婧2, 赵会纳2, 王兵2, 许本波1, 雷波2(
), 夏海乾2, 李永铃1, 张文建3, 余婧2, 赵会纳2, 王兵2, 许本波1, 雷波2( )
)
收稿日期:2024-09-14
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
雷波,女,博士,研究员,研究方向 :烟草分子生物学;E-mail: leibo_1981@163.com作者简介:卢勇杰,男,硕士研究生,研究方向 :烟草分子生物学;E-mail: lyj_luyongjie@163.com
基金资助:
LU Yong-jie1( ), XIA Hai-qian2, LI Yong-ling1, ZHANG Wen-jian3, YU Jing2, ZHAO Hui-na2, WANG Bing2, XU Ben-bo1, LEI Bo2(
), XIA Hai-qian2, LI Yong-ling1, ZHANG Wen-jian3, YU Jing2, ZHAO Hui-na2, WANG Bing2, XU Ben-bo1, LEI Bo2( )
)
Received:2024-09-14
Published:2025-04-26
Online:2025-04-25
摘要:
目的 AP2/ERF(APETALA2/ethylene-responsive factor)转录因子家族中的ESR2基因对植物叶芽发育调控具有重要的作用。克隆烟草NtESR2基因并分析其功能,为烟草叶芽的发育调控提供靶标基因。 方法 采用同源克隆从栽培烟草(Nicotiana tabacum L.)中克隆NtESR2基因,并进行生物信息学、基因表达、激素响应和亚细胞定位分析,同时利用CRISPR/Cas9编辑技术创制NtESR2a和NtESR2b双突变体ntesr2,观察ntesr2突变体的表型。 结果 NtESR2在栽培烟草中具有2个拷贝(NtESR2a和NtESR2b),其CDS长度分别为1 188 bp和1 200 bp,分别编码395和399个氨基酸;NtESR2a蛋白定位于细胞核,NtESR2b蛋白定位于细胞核和细胞膜上,2个蛋白都具有1个AP2结构域,属于ERF亚族,其中NtESR2a来自林烟草,NtESR2b来自绒毛状烟草。NtESR2基因在烟草的根、茎、叶、顶芽、腋芽和花中均有表达,其中在顶芽和腋芽表达较高。通过顺式作用元件分析和外源激素处理,表明该基因受到多种植物激素的调控。ntesr2突变体表现出子叶融合和分生组织缺失的性状,NtCUC2、NtLAS、NtREV、NtYUCCA和NtPIN1基因表达显著下调。 结论 从烟草中克隆得到ERF转录因子NtESR2的2个成员,在烟草顶芽和腋芽中表达较高,受到生长素等多种植物激素的调控,NtESR2通过调控生长素的运输来影响烟草叶芽的形态建立过程,研究结果为调控烟草叶芽发育提供理论依据。
卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277.
LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum[J]. Biotechnology Bulletin, 2025, 41(4): 266-277.
| 引物Primer | 序列Sequence (5′‒3′) | 用途Usage |
|---|---|---|
| NtESR2a-F | ATGGAAGATGCCATGAG | NtESR2a基因克隆 NtESR2a amplification |
| NtESR2a-R | TTAAGAATTCTGCAGCTTAG | |
| NtESR2b-F | ATGCGCGGTGTTAAAGC | NtESR2b基因克隆 NtESR2b amplification |
| NtESR2b-R | TCAAGAATTCTGCAGCTTAG | |
| Actin-Q-F | CGGAATCCACGAGACTACATA | RT-qPCR内标基因检测 RT-qPCR internal standard gene detection |
| Actin-Q-R | GGGAAGCCAAGATAGAGC | |
| ESR2a-Q-F | CCAACTTCCACTAACGATGGGT | RT-qPCR检测NtESR2a基因表达量 RT-qPCR detection of NtESR2a gene expression |
| ESR2a-Q-R | GTGTCTGAAGAAAGGGAACTGGT | |
| ESR2b-Q-F | GTGAACTTCTTAGTTCGAACCCCT | RT-qPCR检测NtESR2b基因表达量 RT-qPCR detection of NtESR2b gene expression |
| ESR2b-Q-R | GACTTGATGGGCTTGGGCT | |
| NtESR2a-CRISPR test-F | CATCTGTTGGGGCGGTGAGGTAT | 检测NtESR2a基因突变 |
| NtESR2a-CRISPR test-R | CTGGTAGTATTGGAGGAATTG | Detection of NtESR2a gene mutation |
| NtESR2b-CRISPR test-F | CATCTGTCGGGGCGGTGAGGTAC | 检测NtESR2b基因突变 |
| NtESR2b-CRISPR test-R | CAAAGAATCATTGCTTTTCTGA | Detection of NtESR2b gene mutation |
| CUC2-Q-F | TCCTCTGTCGGTGCCAATTC | RT-qPCR检测NtCUC2基因表达量 |
| CUC2-Q-R | TTGGGTCGAAACTGTGCGTA | RT-qPCR detection of NtCUC2 gene expression |
| LAS-Q-F | ACAGCTGCAGTAAAGGTGCC | RT-qPCR检测NtLAS基因表达量 |
| LAS-Q-R | GCGAGGATACTCTCCCAGCA | RT-qPCR detection of NtLAS gene expression |
| REV-Q-F | CCACTGGGCCACACAATAGAA | RT-qPCR检测NtREV基因表达量 |
| REV-Q-R | ACAGATCGTGCACTGTAGCC | RT-qPCR detection of NtREV gene expression |
| YUCCA-Q-F | TGCTAAGTACGACGAAGCTTGT | RT-qPCR检测NtYUCCA基因表达量 |
| YUCCA-Q-R | ATGCCAGAATTTCCACAGCCTA | RT-qPCR detection of NtYUCCA gene expression |
| PIN1-Q-F | TCGAACCAAGTGAGGGCATC | RT-qPCR检测NtPIN1基因表达量 |
| PIN1-Q-R | AGCTGCAATCAGAGAAGCGA | RT-qPCR detection of NtPIN1 gene expression |
表1 引物序列
Table 1 Primer Sequence
| 引物Primer | 序列Sequence (5′‒3′) | 用途Usage |
|---|---|---|
| NtESR2a-F | ATGGAAGATGCCATGAG | NtESR2a基因克隆 NtESR2a amplification |
| NtESR2a-R | TTAAGAATTCTGCAGCTTAG | |
| NtESR2b-F | ATGCGCGGTGTTAAAGC | NtESR2b基因克隆 NtESR2b amplification |
| NtESR2b-R | TCAAGAATTCTGCAGCTTAG | |
| Actin-Q-F | CGGAATCCACGAGACTACATA | RT-qPCR内标基因检测 RT-qPCR internal standard gene detection |
| Actin-Q-R | GGGAAGCCAAGATAGAGC | |
| ESR2a-Q-F | CCAACTTCCACTAACGATGGGT | RT-qPCR检测NtESR2a基因表达量 RT-qPCR detection of NtESR2a gene expression |
| ESR2a-Q-R | GTGTCTGAAGAAAGGGAACTGGT | |
| ESR2b-Q-F | GTGAACTTCTTAGTTCGAACCCCT | RT-qPCR检测NtESR2b基因表达量 RT-qPCR detection of NtESR2b gene expression |
| ESR2b-Q-R | GACTTGATGGGCTTGGGCT | |
| NtESR2a-CRISPR test-F | CATCTGTTGGGGCGGTGAGGTAT | 检测NtESR2a基因突变 |
| NtESR2a-CRISPR test-R | CTGGTAGTATTGGAGGAATTG | Detection of NtESR2a gene mutation |
| NtESR2b-CRISPR test-F | CATCTGTCGGGGCGGTGAGGTAC | 检测NtESR2b基因突变 |
| NtESR2b-CRISPR test-R | CAAAGAATCATTGCTTTTCTGA | Detection of NtESR2b gene mutation |
| CUC2-Q-F | TCCTCTGTCGGTGCCAATTC | RT-qPCR检测NtCUC2基因表达量 |
| CUC2-Q-R | TTGGGTCGAAACTGTGCGTA | RT-qPCR detection of NtCUC2 gene expression |
| LAS-Q-F | ACAGCTGCAGTAAAGGTGCC | RT-qPCR检测NtLAS基因表达量 |
| LAS-Q-R | GCGAGGATACTCTCCCAGCA | RT-qPCR detection of NtLAS gene expression |
| REV-Q-F | CCACTGGGCCACACAATAGAA | RT-qPCR检测NtREV基因表达量 |
| REV-Q-R | ACAGATCGTGCACTGTAGCC | RT-qPCR detection of NtREV gene expression |
| YUCCA-Q-F | TGCTAAGTACGACGAAGCTTGT | RT-qPCR检测NtYUCCA基因表达量 |
| YUCCA-Q-R | ATGCCAGAATTTCCACAGCCTA | RT-qPCR detection of NtYUCCA gene expression |
| PIN1-Q-F | TCGAACCAAGTGAGGGCATC | RT-qPCR检测NtPIN1基因表达量 |
| PIN1-Q-R | AGCTGCAATCAGAGAAGCGA | RT-qPCR detection of NtPIN1 gene expression |
蛋白 Protein | 分子式 Formula | 分子量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 总平均亲水性系数 GRAVY |
|---|---|---|---|---|---|
| NtESR2a | C1871H2899N533O611S18 | 43.2 | 7.59 | 61.93 | -0.557 |
| NtESR2b | C1895H2940N534O620S18 | 43.7 | 6.95 | 56.45 | -0.562 |
表2 NtESR2蛋白的理化性质分析
Table 2 Analysis of the physicochemical properties of NtESR2 proteins
蛋白 Protein | 分子式 Formula | 分子量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 总平均亲水性系数 GRAVY |
|---|---|---|---|---|---|
| NtESR2a | C1871H2899N533O611S18 | 43.2 | 7.59 | 61.93 | -0.557 |
| NtESR2b | C1895H2940N534O620S18 | 43.7 | 6.95 | 56.45 | -0.562 |

图3 ESR2蛋白序列比对(A)、系统进化树构建及motif分析(B)NsESR2:林烟草,XP_009765569.1;NtomESR2:绒毛状烟草,XP_009598275.1;SlLFS:番茄,XP_004239483.2;StESR2:马铃薯,XP_006357626.1;ApESR2:穿心莲,XP_051122507.1;HbESR2:橡胶树,XP_021657412.1;AtESR2:拟南芥,NP_173864.1;BnESR2:欧洲油菜,XP_013726908.1;ZmESR2:玉米,XP_008678756.1;ObESR2:短花稻,XP_040377266.1
Fig. 3 ESR2 protein sequence alignment (A), phylogenetic tree construction and motif analysis (B)NsESR2: Nicotiana sylvestris, XP_009765569.1; NtomESR2: Nicotiana tomentosiformis, XP_009598275.1; SlLFS: Solanum lycopersicum, XP_004239483. 2; StESR2: Solanum tuberosum, XP_006357626. 1; ApESR2: Andrographis paniculata, XP_051122507.1; HbESR2: Hevea brasiliensis, XP_021657412.1; AtESR2: Arabidopsis thaliana, NP_173864.1; BnESR2: Brassica napus, XP_013726908.1; ZmESR2: Zea mays, XP_008678756.1; ObESR2: Oryza brachyantha, XP_040377266.1

图5 NtESR2基因在不同组织中的表达分析不同小写字母表示差异达到显著水平(P<0.05)。下同
Fig. 5 Expression analysis of NtESR2 gene in different tissuesDifferent lower letters indicate significant difference (P<0.05). The same below

图9 突变体材料和野生型材料表型观察A,C:野生型WT;B,D:ntesr2突变体
Fig. 9 Phenotypic observation of mutant materials and wild-type materialsA, C: Wild type WT; B, D: ntesr2 double copy mutant
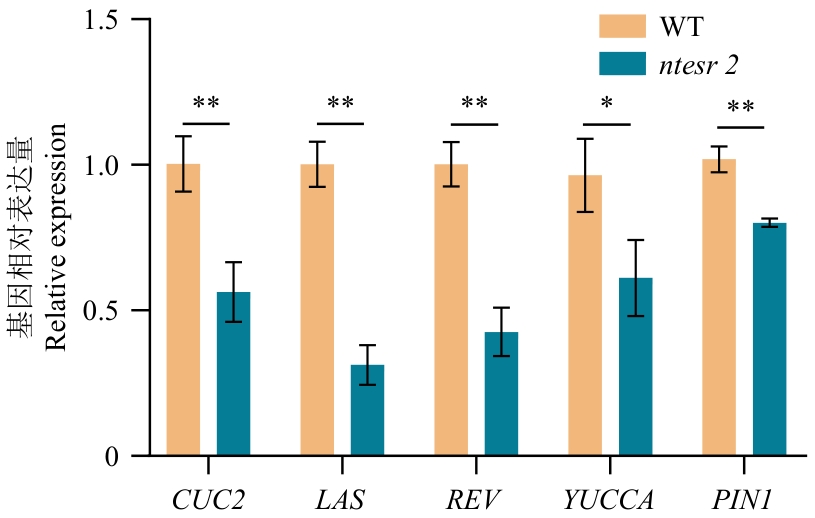
图10 ntesr2突变体与野生型WT的基因表达分析*表示显著性P<0.05,**表示显著性P<0.01
Fig. 10 Analysis of gene expression in ntesr2 mutant and wild‑type* indicates significance P<0.05, and ** indicates significance P<0.01
| 1 | Feng K, Hou XL, Xing GM, et al. Advances in AP2/ERF super-family transcription factors in plant [J]. Crit Rev Biotechnol, 2020, 40(6): 750-776. |
| 2 | Xie ZL, Nolan T, Jiang H, et al. The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis [J]. Plant Cell, 2019, 31(8): 1788-1806. |
| 3 | Owji H, Hajiebrahimi A, Seradj H, et al. Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis [J]. Comput Biol Chem, 2017, 71: 32-56. |
| 4 | Ghorbani R, Zakipour Z, Alemzadeh A, et al. Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus [J]. Physiol Mol Biol Plants, 2020, 26(7): 1463-1476. |
| 5 | Chen K, Tang WS, Zhou YB, et al. AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs [J]. Plant Physiol Biochem, 2022, 170: 287-295. |
| 6 | Kirch T, Simon R, Grünewald M, et al. The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development [J]. Plant Cell, 2003, 15(3): 694-705. |
| 7 | Marsch-Martinez N, Greco R, Becker JD, et al. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways [J]. Plant Mol Biol, 2006, 62(6): 825-843. |
| 8 | Nag A, Yang YZ, Jack T. DORNROSCHEN-LIKE an AP2 gene, is necessary for stamen emergence in Arabidopsis [J]. Plant Mol Biol, 2007, 65(3): 219-232. |
| 9 | Ikeda Y, Králová M, Zalabák D, et al. Post-embryonic lateral organ development and adaxial-abaxial polarity are regulated by the combined effect of ENHANCER OF SHOOT REGENERATION 1 and WUSCHEL in Arabidopsis shoots [J]. Int J Mol Sci, 2021, 22(19): 10621. |
| 10 | Capua Y, Eshed Y. Coordination of auxin-triggered leaf initiation by tomato LEAFLESS [J]. Proc Natl Acad Sci U S A, 2017, 114(12): 3246-3251. |
| 11 | Lu R, Hu SQ, Feng J, et al. The AP2 transcription factor BARE RECEPTACLE regulates floral organogenesis via auxin pathways in woodland strawberry [J]. Plant Cell, 2024, 36(12): 4970-4987. |
| 12 | Long JA, Moan EI, Medford JI, et al. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis [J]. Nature, 1996, 379(6560): 66-69. |
| 13 | 王兵, 赵会纳, 余婧, 等. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控 [J]. 生物技术通报, 2023, 39(10): 197-208. |
| Wang B, Zhao HN, Yu J, et al. Regulation of leaf bud by REVOLUTA in tobacco based on CRISPR/Cas9 system [J]. Biotechnol Bull, 2023, 39(10): 197-208. | |
| 14 | Zhang C, Wang J, Wenkel S, et al. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators [J]. Development, 2018, 145(24): dev158352. |
| 15 | Greb T, Clarenz O, Schafer E, et al. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation [J]. Genes Dev, 2003, 17(9): 1175-1187. |
| 16 | Hibara KI, Karim MR, Takada S, et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation [J]. Plant Cell, 2006, 18(11): 2946-2957. |
| 17 | Tian CH, Zhang XN, He J, et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation [J]. Mol Syst Biol, 2014, 10(10): 755. |
| 18 | Yamada M, Tanaka S, Miyazaki T, et al. Expression of the auxin biosynthetic genes YUCCA1 and YUCCA4 is dependent on the boundary regulators CUP-SHAPED COTYLEDON genes in the Arabidopsis thaliana embryo [J]. Plant Biotechnol, 2022, 39(1): 37-42. |
| 19 | Lee BH, Jeon JO, Lee MM, et al. Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation [J]. Plant Signal Behav, 2015, 10(2): e988071. |
| 20 | Scofield S, Murison A, Jones A, et al. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network [J]. Development, 2018, 145(9): dev157081. |
| 21 | Spinelli SV, Martin AP, Viola IL, et al. A mechanistic link between STM and CUC1 during Arabidopsis development [J]. Plant Physiol, 2011, 156(4): 1894-1904. |
| 22 | Keller T, Abbott J, Moritz T, et al. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development [J]. Plant Cell, 2006, 18(3): 598-611. |
| 23 | Yang F, Wang Q, Schmitz G, et al. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis [J]. Plant J, 2012, 71(1): 61-70. |
| 24 | Leitch IJ, Hanson L, Lim KY, et al. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) [J]. Ann Bot, 2008, 101(6): 805-814. |
| 25 | Matsuo N, Makino M, Banno H. Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated [J]. Plant Sci, 2011, 181(1): 39-46. |
| 26 | Möller B, Weijers D. Auxin control of embryo patterning [J]. Cold Spring Harb Perspect Biol, 2009, 1(5): a001545. |
| 27 | Brandt R, Salla-Martret M, Bou-Torrent J, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses [J]. Plant J, 2012, 72(1): 31-42. |
| 28 | O'Connor DL, Runions A, Sluis A, et al. A division in PIN-mediated auxin patterning during organ initiation in grasses [J]. PLoS Comput Biol, 2014, 10(1): e1003447. |
| 29 | Gälweiler L, Guan C, Müller A, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue [J]. Science, 1998, 282(5397): 2226-2230. |
| 30 | Cheng YF, Dai XH, Zhao YD. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis [J]. Plant Cell, 2007, 19(8): 2430-2439. |
| 31 | Cao X, Yang HL, Shang CQ, et al. The roles of auxin biosynthesis YUCCA gene family in plants [J]. Int J Mol Sci, 2019, 20(24): 6343. |
| [1] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [2] | 班秋艳, 赵鑫月, 迟文静, 黎俊生, 王琼, 夏瑶, 梁丽云, 贺巍, 李叶云, 赵广山. 茶树光敏色素互作因子CsPIF3a的克隆及其与光温逆境的响应[J]. 生物技术通报, 2025, 41(4): 256-265. |
| [3] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [4] | 王琛, 刘国梅, 陈畅, 张晋龙, 姚琳, 孙璇, 杜春芳. 白菜型油菜CCDs家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(3): 161-170. |
| [5] | 马耀武, 张麒宇, 杨淼, 蒋诚, 张振宇, 张伊琳, 李梦莎, 许嘉阳, 张斌, 崔光周, 姜瑛. 烟草根际促生菌的筛选鉴定及促生性能研究[J]. 生物技术通报, 2025, 41(3): 271-281. |
| [6] | 韩梦荞, 吴疆, 李丽华, 王昭懿, 邓习, 韦凤杰, 任民, 孙洋洋, 李富欣. 烟草染色体制备体系的建立与优化[J]. 生物技术通报, 2025, 41(3): 44-50. |
| [7] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [8] | 李明, 刘祥宇, 王益娜, 和四梅, 沙本才. 紫金龙异紫堇定生物合成相关6-OMT基因克隆与功能表征[J]. 生物技术通报, 2025, 41(2): 309-320. |
| [9] | 王子傲, 田瑞, 崔永梅, 白羿雄, 姚晓华, 安立昆, 吴昆仑. 青稞HvnJAZ4的生物信息学和表达模式分析[J]. 生物技术通报, 2025, 41(1): 173-185. |
| [10] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [11] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [12] | 崔海洋, 谭淼, 全壮, 陈红利, 董艳敏, 唐立春. 利用Cas9TX实现非病毒TRAC定点整合制备T细胞[J]. 生物技术通报, 2024, 40(9): 190-197. |
| [13] | 张曼玉, 董嘉诚, 苟福凡, 弓朝晖, 刘倩, 孙文良, 孔臻, 郝捷, 王敏, 田朝光. 嗜热毁丝霉果胶酯酶MtCE12-1的克隆表达及其酶学性质和应用研究[J]. 生物技术通报, 2024, 40(9): 291-300. |
| [14] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [15] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||