








生物技术通报 ›› 2025, Vol. 41 ›› Issue (11): 75-88.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0153
吕欢欢1( ), 张高阳1(
), 张高阳1( ), 王赛笛1, 孙忠科1, 李成伟1, 罗德平2(
), 王赛笛1, 孙忠科1, 李成伟1, 罗德平2( )
)
收稿日期:2025-01-20
出版日期:2025-11-26
发布日期:2025-12-09
通讯作者:
张高阳,男,博士,讲师,研究方向 :植物遗传合成;E-mail: gaoyangzhang@haut.edu.cn作者简介:吕欢欢,女,硕士研究生,研究方向 :代谢产物合成;E-mail: lvhuanhuan@stu.haut.edu.cn
基金资助:
LYU Huan-huan1( ), ZHANG Gao-yang1(
), ZHANG Gao-yang1( ), WANG Sai-di1, SUN Zhong-ke1, LI Cheng-wei1, LUO De-ping2(
), WANG Sai-di1, SUN Zhong-ke1, LI Cheng-wei1, LUO De-ping2( )
)
Received:2025-01-20
Published:2025-11-26
Online:2025-12-09
摘要:
5-氨基乙酰丙酸(5-ALA)是生物体内天然存在一种非蛋白质类氨基酸,目前已被广泛应用于医药、保健、农业和动物添加剂等领域。5-ALA主要通过化学和生物法合成,但由于化学合成的工艺复杂性和成本问题限制了其规模化发展,而生物合成法以其环保、高效等优势彰显了其工业化发展的巨大潜力。近年来,由于合成生物学和代谢工程等交叉学科的迅猛发展,5-ALA的生物合成已逐渐成为研究热点。本文回溯了过去几十年5-ALA生物合成的发展历程,总结并梳理了基于C4和C5途径、辅因子再生途径、三羧酸循环等途径、用于高效细胞工厂构建的各种代谢工程策略的研究进展,重点阐述了基于生物传感器的动态调控和高通量筛选方法的建立等策略在菌株代谢工程改造中的应用和重要性,并就进一步提高5-ALA的合成产量,打破其工业化应用的瓶颈提出了几点策略和展望,以期为5-ALA的高效合成和产业制造提供一定的参考和研究依据。
吕欢欢, 张高阳, 王赛笛, 孙忠科, 李成伟, 罗德平. 5-氨基乙酰丙酸(5-ALA)的生物合成研究进展[J]. 生物技术通报, 2025, 41(11): 75-88.
LYU Huan-huan, ZHANG Gao-yang, WANG Sai-di, SUN Zhong-ke, LI Cheng-wei, LUO De-ping. Advances in the Biosynthesis of 5-aminolevulinic Acid (5-ALA)[J]. Biotechnology Bulletin, 2025, 41(11): 75-88.
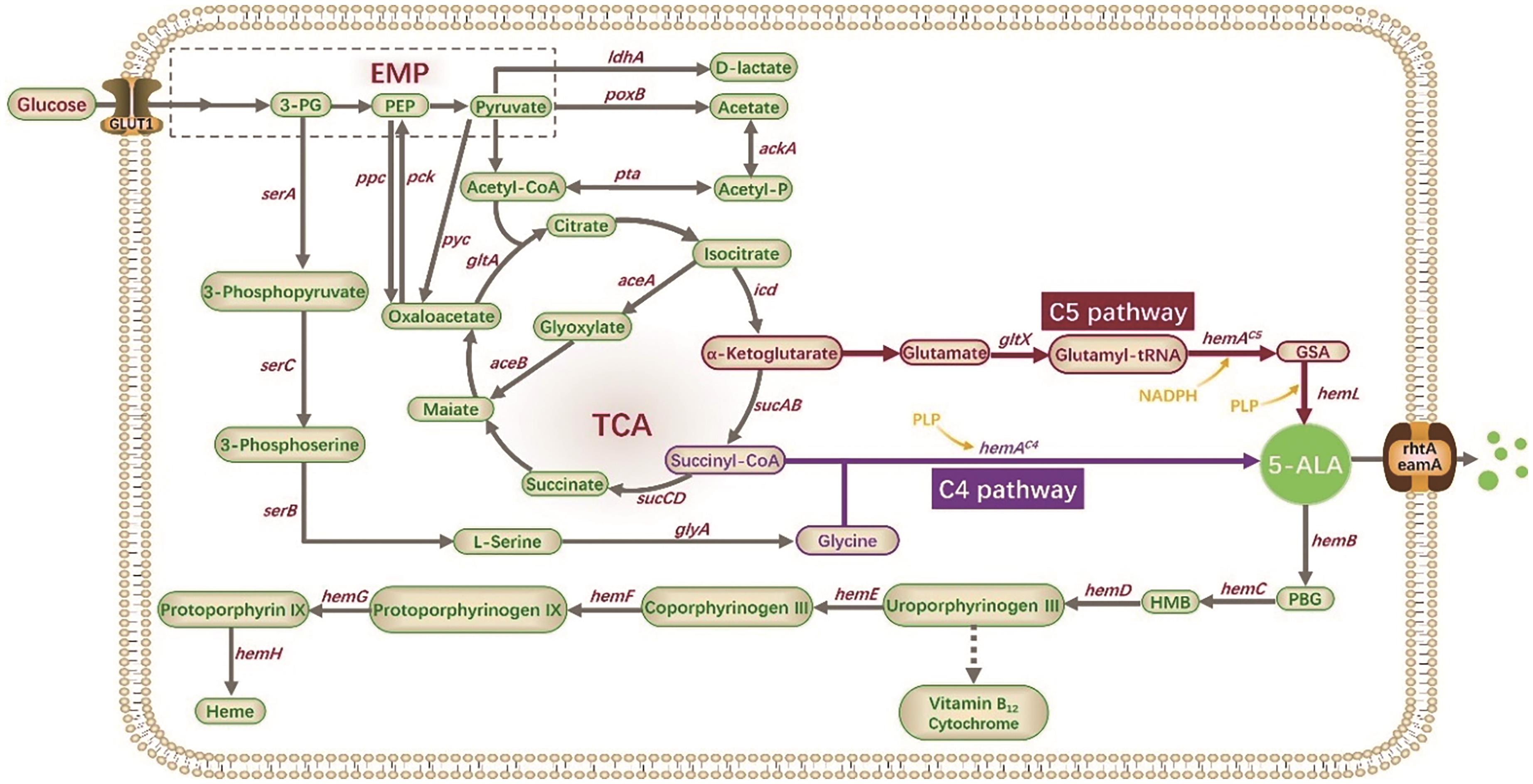
图2 5-ALA及四吡咯化合物的生物合成途径3-PG: 3-phosphoglycerate; PEP: phosphoenolpyruvate; GSA: glutamate-1-semialdehyde; 5-ALA: 5-aminolevulinic acid; PBG: porphobilinogen; HMB: hydroxymethylbilane
Fig. 2 Biosynthetic pathways of 5-ALA and tetrapyrrole compounds
菌株 Strain | 底物 Substrate | 策略 Strategy | 滴度 Titer (g/L) | 参考文献 Reference |
|---|---|---|---|---|
大肠杆菌 E. coli | 葡萄糖Glucose 甘油Glycerol | 过表达荚膜红细菌来源的ALAS酶并进行密码子优化,过表达伴侣蛋白GroELS,培养基中添加铁离子 | 15.6 | [ |
葡萄糖Glucose 谷氨酸Glutamate | 异源表达拟南芥来源的hemA基因和GluTR调节蛋白PGR7,优化宿主菌株 | 7.64 | [ | |
| 葡萄糖Glucose | 选择不同来源的C5途径关键酶并优化其表达比例,微调竞争性代谢途径,优化辅因子再生途径 | 12.1 | [ | |
大肠杆菌 E. coliB | 葡萄糖Glucose 甘氨酸Glycine | 过表达荚膜红细菌来源的ALAS酶,加强PLP补救途径 | 8.21 | [ |
葡萄糖Glucose 甘氨酸Glycine琥珀酸Succinate | 过表达球形红杆菌来源的hemA基因,过表达外排蛋白基因rhtA, 过表达pdxK和pdxY基因以增加辅因子PLP的再生,引入伴侣蛋白DnaK和GroELS | 7.47 | [ | |
| 葡萄糖Glucose | 过表达ALAS酶,过表达过氧化氢酶CAT和超氧化物歧化酶SOD以增强抗氧化系统 | 11.5 | [ | |
葡萄糖Glucose 磷酸吡哆醛PLP 泛酸PA | 进化分析筛选新的ALAS酶,识别并修改4个关键靶标,开发动态控制系统以平衡氧化还原稳态和碳通量,协同优化辅因子 | 63.39 | [ | |
| 葡萄糖Glucose | 过表达hemARS 基因,基于RGMS技术的eamA基因定向进化,过表达sodB和katE基因提高抗氧化性,减弱hemB基因的表达 | 19.02 | [ | |
葡萄糖Glucose 甘氨酸Glycine | 建立5-ALA响应生物传感器,筛选HemA酶和菌株突变体,加强5-ALA外排,抑制下游基因表达,提高前体和辅因子供应 | 58.54 | [ | |
谷氨酸棒状杆菌 C. glutamicum | 葡萄糖Glucose | 过表达荚膜红细菌来源的hemA基因,过表达外排蛋白基因rhtA,敲除sucCD基因,采用“生长-产酸”两阶段发酵模式 | 14.7 | [ |
| 葡萄糖Glucose | RBS工程优化RphemA基因的表达,敲除odhI和sdhA基因,用small RNA抑制hemB基因的表达,过表达外排蛋白基因eamA | 25.05 | [ | |
| 葡萄糖Glucose | 定向诱变odhI基因,增加谷氨酸供应,添加诱导剂吐温40和乙胺丁醇 | 2.9 | [ | |
葡萄糖Glucose 甘氨酸Glycine | 过表达球形红杆菌来源的ALAS酶,削弱竞争途径,过表达ppc基因和外排蛋白基因rhtA,敲除细胞壁合成相关蛋白HMW-PBP | 7.53 | [ | |
葡萄糖Glucose 木薯渣Cassava bagasse | 筛选比较不同来源的ALAS酶,RBS工程优化hemA基因和ppc基因平衡表达,利用廉价可再生原料替代葡萄糖 | 18.5 | [ | |
| 葡萄糖Glucose | 通过3种不同策略调节odhA基因表达,响应血红素浓度变化自动调节rhtA基因表达,精确调控5-ALA合成和辅因子再生关键基因 | 3.16 | [ |
表1 代谢工程改造微生物合成5-ALA的进展
Table 1 Progress in metabolic engineering for microbial synthesis of 5-ALA
菌株 Strain | 底物 Substrate | 策略 Strategy | 滴度 Titer (g/L) | 参考文献 Reference |
|---|---|---|---|---|
大肠杆菌 E. coli | 葡萄糖Glucose 甘油Glycerol | 过表达荚膜红细菌来源的ALAS酶并进行密码子优化,过表达伴侣蛋白GroELS,培养基中添加铁离子 | 15.6 | [ |
葡萄糖Glucose 谷氨酸Glutamate | 异源表达拟南芥来源的hemA基因和GluTR调节蛋白PGR7,优化宿主菌株 | 7.64 | [ | |
| 葡萄糖Glucose | 选择不同来源的C5途径关键酶并优化其表达比例,微调竞争性代谢途径,优化辅因子再生途径 | 12.1 | [ | |
大肠杆菌 E. coliB | 葡萄糖Glucose 甘氨酸Glycine | 过表达荚膜红细菌来源的ALAS酶,加强PLP补救途径 | 8.21 | [ |
葡萄糖Glucose 甘氨酸Glycine琥珀酸Succinate | 过表达球形红杆菌来源的hemA基因,过表达外排蛋白基因rhtA, 过表达pdxK和pdxY基因以增加辅因子PLP的再生,引入伴侣蛋白DnaK和GroELS | 7.47 | [ | |
| 葡萄糖Glucose | 过表达ALAS酶,过表达过氧化氢酶CAT和超氧化物歧化酶SOD以增强抗氧化系统 | 11.5 | [ | |
葡萄糖Glucose 磷酸吡哆醛PLP 泛酸PA | 进化分析筛选新的ALAS酶,识别并修改4个关键靶标,开发动态控制系统以平衡氧化还原稳态和碳通量,协同优化辅因子 | 63.39 | [ | |
| 葡萄糖Glucose | 过表达hemARS 基因,基于RGMS技术的eamA基因定向进化,过表达sodB和katE基因提高抗氧化性,减弱hemB基因的表达 | 19.02 | [ | |
葡萄糖Glucose 甘氨酸Glycine | 建立5-ALA响应生物传感器,筛选HemA酶和菌株突变体,加强5-ALA外排,抑制下游基因表达,提高前体和辅因子供应 | 58.54 | [ | |
谷氨酸棒状杆菌 C. glutamicum | 葡萄糖Glucose | 过表达荚膜红细菌来源的hemA基因,过表达外排蛋白基因rhtA,敲除sucCD基因,采用“生长-产酸”两阶段发酵模式 | 14.7 | [ |
| 葡萄糖Glucose | RBS工程优化RphemA基因的表达,敲除odhI和sdhA基因,用small RNA抑制hemB基因的表达,过表达外排蛋白基因eamA | 25.05 | [ | |
| 葡萄糖Glucose | 定向诱变odhI基因,增加谷氨酸供应,添加诱导剂吐温40和乙胺丁醇 | 2.9 | [ | |
葡萄糖Glucose 甘氨酸Glycine | 过表达球形红杆菌来源的ALAS酶,削弱竞争途径,过表达ppc基因和外排蛋白基因rhtA,敲除细胞壁合成相关蛋白HMW-PBP | 7.53 | [ | |
葡萄糖Glucose 木薯渣Cassava bagasse | 筛选比较不同来源的ALAS酶,RBS工程优化hemA基因和ppc基因平衡表达,利用廉价可再生原料替代葡萄糖 | 18.5 | [ | |
| 葡萄糖Glucose | 通过3种不同策略调节odhA基因表达,响应血红素浓度变化自动调节rhtA基因表达,精确调控5-ALA合成和辅因子再生关键基因 | 3.16 | [ |
| [1] | Kang Z, Zhang JL, Zhou JW, et al. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12 [J]. Biotechnol Adv, 2012, 30(6): 1533-1542. |
| [2] | Yi YC, Shih IT, Yu TH, et al. Challenges and opportunities of bioprocessing 5-aminolevulinic acid using genetic and metabolic engineering: a critical review [J]. Bioresour Bioprocess, 2021, 8(1): 100. |
| [3] | Pignatelli P, Umme S, D’Antonio DL, et al. Reactive oxygen species produced by 5-aminolevulinic acid photodynamic therapy in the treatment of cancer [J]. Int J Mol Sci, 2023, 24(10): 8964. |
| [4] | Higashikawa F, Kanno K, Ogata A, et al. Reduction of fatigue and anger-hostility by the oral administration of 5-aminolevulinic acid phosphate: a randomized, double-blind, placebo-controlled, parallel study [J]. Sci Rep, 2020, 10(1): 16004. |
| [5] | Yang LY, Deng HH, Chen YM, et al. 5-aminolevulinic acid-hyaluronic acid complexes enhance skin retention of 5-aminolevulinic acid and therapeutic efficacy in the treatment of hypertrophic scar [J]. AAPS PharmSciTech, 2022, 23(6): 216. |
| [6] | Tan SY, Cao J, Xia XL, et al. Advances in 5-aminolevulinic acid priming to enhance plant tolerance to abiotic stress [J]. Int J Mol Sci, 2022, 23(2): 702. |
| [7] | Cheng FX, Wang J, Song ZQ, et al. Nematicidal effects of 5-aminolevulinic acid on plant-parasitic nematodes [J]. J Nematol, 2017, 49(3): 295-303. |
| [8] | Hendawy AO, Khattab MS, Sugimura S, et al. Effects of 5-aminolevulinic acid as a supplement on animal performance, iron status, and immune response in farm animals: a review [J]. Animals, 2020, 10(8): 1352. |
| [9] | Chen YY, Huang JC, Wu CY, et al. A comprehensive review on the recent advances for 5-aminolevulinic acid production by the engineered bacteria [J]. Crit Rev Biotechnol, 2025, 45(1): 148-163. |
| [10] | QYResearch(恒州博智). 2025-2031全球及中国5-ALA行业研究及十五五规划分析报告 [EB]. (2025-03-19). |
| QYResearch, 2025-2031 Global and China 5-ALA Industry Research and 15th Five-Year Plan Analysis Report [EB]. (2025-03-19). | |
| [11] | Zhou HM, Zhang CY, Li ZL, et al. Systematic development of a highly efficient cell factory for 5-aminolevulinic acid production [J]. Trends Biotechnol, 2024, 42(11): 1479-1502. |
| [12] | Luo ZS, Yu SQ, Zeng WZ, et al. Comparative analysis of the chemical and biochemical synthesis of keto acids [J]. Biotechnol Adv, 2021, 47: 107706. |
| [13] | Jiang MR, Hong KQ, Mao YF, et al. Natural 5-aminolevulinic acid: sources, biosynthesis, detection and applications [J]. Front Bioeng Biotechnol, 2022, 10: 841443. |
| [14] | Kang Z, Ding WW, Gong X, et al. Recent advances in production of 5-aminolevulinic acid using biological strategies [J]. World J Microbiol Biotechnol, 2017, 33(11): 200. |
| [15] | Fu WQ, Lin JP, Cen PL. Expression of a hemA gene from Agrobacterium radiobacter in a rare codon optimizing Escherichia coli for improving 5-aminolevulinate production [J]. Appl Biochem Biotechnol, 2010, 160(2): 456-466. |
| [16] | Kang DK, Kim SS, Chi WJ, et al. Cloning and expression of the Rhodobacter capsulatus hemA gene in E. coli for the production of 5-aminolevulinic acid [J]. Journal of Microbiology and Biotechnology, 2004, 14(6): 1327-1332. |
| [17] | Shemin D, Russell CS. δ-aminolevulinic acid, its role in the biosynthesis of porphyrins and PURINES [J]. J Am Chem Soc, 1953, 75(19): 4873-4874. |
| [18] | Ge FL, Li XK, Ge QR, et al. Modular control of multiple pathways of Corynebacterium glutamicum for 5-aminolevulinic acid production [J]. AMB Express, 2021, 11(1): 179. |
| [19] | Zou YL, Chen T, Feng LL, et al. Enhancement of 5-aminolevulinic acid production by metabolic engineering of the Glycine biosynthesis pathway in Corynebacterium glutamicum [J]. Biotechnol Lett, 2017, 39(9): 1369-1374. |
| [20] | Yang P, Liu WJ, Cheng XL, et al. A new strategy for production of 5-aminolevulinic acid in recombinant Corynebacterium glutamicum with high yield [J]. Appl Environ Microbiol, 2016, 82(9): 2709-2717. |
| [21] | Sasaki K, Watanabe M, Tanaka T, et al. Biosynthesis, biotechnological production and applications of 5-aminolevulinic acid [J]. Appl Microbiol Biotechnol, 2002, 58(1): 23-29. |
| [22] | Liu SL, Zhang GM, Li XK, et al. Microbial production and applications of 5-aminolevulinic acid [J]. Appl Microbiol Biotechnol, 2014, 98(17): 7349-7357. |
| [23] | Stojanovski BM, Hunter GA, Jahn M, et al. Unstable reaction intermediates and hysteresis during the catalytic cycle of 5-aminolevulinate synthase implications from using pseudo and alternate substrates and a promiscuous enzyme variant [J]. J Biol Chem, 2014, 289(33): 22915-22925. |
| [24] | Beale SI. The biosynthesis of delta-aminolevulinic acid in Chlorella [J]. Plant Physiol, 1970, 45(4): 504-506. |
| [25] | Schneegurt MA, Beale SI. Characterization of the RNA required for biosynthesis of δ-aminolevulinic acid from glutamate [J]. Plant Physiol, 1988, 86(2): 497-504. |
| [26] | Li JM, Russell CS, Cosloy SD. Cloning and structure of the hemA gene of Escherichia coli K-12 [J]. Gene, 1989, 82(2): 209-217. |
| [27] | Schauer S, Chaturvedi S, Randau L, et al. Escherichia coli Glutamyl-tRNA Reductase trapping the thioester intermediate [J]. J Biol Chem, 2002, 277(50): 48657-48663. |
| [28] | Jahn D, Verkamp E, Söll D. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis [J]. Trends Biochem Sci, 1992, 17(6): 215-218. |
| [29] | Zhang JL, Kang Z, Chen J, et al. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli [J]. Sci Rep, 2015, 5: 8584. |
| [30] | Weinstein JD, Beale SI. Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis [J]. J Biol Chem, 1983, 258(11): 6799-6807. |
| [31] | Pedró-Rosa L, Buckner FS, Ranade RM, et al. Identification of potent inhibitors of the Trypanosoma brucei methionyl-tRNA synthetase via high-throughput orthogonal screening [J]. SLAS Discov, 2015, 20(1): 122-130. |
| [32] | Ge JZ, Wang XL, Bai YG, et al. Engineering Escherichia coli for efficient assembly of heme proteins [J]. Microb Cell Fact, 2023, 22(1): 59. |
| [33] | Lüer C, Schauer S, Möbius K, et al. Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2, 1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis [J]. J Biol Chem, 2005, 280(19): 18568-18572. |
| [34] | Wang P, Liang FC, Wittmann D, et al. Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis [J]. Proc Natl Acad Sci USA, 2018, 115(15): E3588-E3596. |
| [35] | Chen HX, Jiang P. Metabolic engineering of Escherichia coli for efficient biosynthesis of fluorescent phycobiliprotein [J]. Microb Cell Fact, 2019, 18(1): 58. |
| [36] | Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens [J]. J Mol Biol, 2016, 428(17): 3408-3428. |
| [37] | Sasaki K, Ikeda S, Nishizawa Y, et al. Production of 5-aminolevulinic acid by photosynthetic bacteria [J]. J Ferment Technol, 1987, 65(5): 511-515. |
| [38] | Nishikawa S, Watanabe K, Tanaka T, et al. Rhodobacter sphaeroides mutants which accumulate 5-aminolevulinic acid under aerobic and dark conditions [J]. J Biosci Bioeng, 1999, 87(6): 798-804. |
| [39] | Yu TH, Tan SI, Yi YC, et al. New insight into the codon usage and medium optimization toward stable and high-level 5-aminolevulinic acid production in Escherichia coli [J]. Biochem Eng J, 2022, 177: 108259. |
| [40] | Zhao AG, Zhai MZ. Production of 5-aminolevulinic acid from glutamate by overexpressing HemA1 and pgr7 from Arabidopsis thaliana in Escherichia coli [J]. World J Microbiol Biotechnol, 2019, 35(11): 175. |
| [41] | Luo ZS, Pan F, Zhu YF, et al. Synergistic improvement of 5-aminolevulinic acid production with synthetic scaffolds and system pathway engineering [J]. ACS Synth Biol, 2022, 11(8): 2766-2778. |
| [42] | Xue CF, Yu TH, Ng IS. Engineering pyridoxal kinase Pd x Y-integrated Escherichia coli strain and optimization for high-level 5-aminolevulinic acid production [J]. J Taiwan Inst Chem Eng, 2021, 120: 49-58. |
| [43] | Shih IT, Yi YC, Ng IS. Plasmid-free system and modular design for efficient 5-aminolevulinic acid production by engineered Escherichia coli [J]. Appl Biochem Biotechnol, 2021, 193(9): 2858-2871. |
| [44] | Zhu CC, Chen JZ, Wang Y, et al. Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli [J]. Biotechnol Bioeng, 2019, 116(8): 2018-2028. |
| [45] | Yang YT, Zou YH, Chen X, et al. Metabolic engineering of Escherichia coli for the production of 5-aminolevulinic acid based on combined metabolic pathway modification and reporter-guided mutant selection (RGMS) [J]. Biotechnol Biofuels Bioprod, 2024, 17(1): 82. |
| [46] | Wang Q, Jia MJ, Li HJ, et al. Design of a genetically encoded biosensor for high-throughput screening and engineering 5-aminolevulinic acid hyper-producing Escherichia coli [J]. ACS Sustainable Chem Eng, 2024, 12(12): 4846-4857. |
| [47] | 王丽君, 闫思翰, 杨套伟, 等. 代谢改造重组谷氨酸棒杆菌C4途径高效合成5-氨基乙酰丙酸 [J]. 生物工程学报, 2021, 37(12): 4314-4328. |
| Wang LJ, Yan SH, Yang TW, et al. Engineering the C4 pathway of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid [J]. Chin J Biotech, 2021, 37(12): 4314-4328. | |
| [48] | Ko YJ, You SK, Kim M, et al. Enhanced production of 5-aminolevulinic acid via flux redistribution of TCA cycle toward l-glutamate in Corynebacterium glutamicum [J]. Biotechnol Bioprocess Eng, 2019, 24(6): 915-923. |
| [49] | Feng LL, Zhang Y, Fu J, et al. Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid [J]. Biotechnol Bioeng, 2016, 113(6): 1284-1293. |
| [50] | Chen JZ, Wang Y, Guo X, et al. Efficient bioproduction of 5-aminolevulinic acid, a promising biostimulant and nutrient, from renewable bioresources by engineered Corynebacterium glutamicum [J]. Biotechnol Biofuels, 2020, 13: 41. |
| [51] | Zhang CL, Li YJ, Zhu FZ, et al. Metabolic engineering of an auto-regulated Corynebacterium glutamicum chassis for biosynthesis of 5-aminolevulinic acid [J]. Bioresour Technol, 2020, 318: 124064. |
| [52] | Guo YY, Zhao HL, Lin ZB, et al. Heme in cardiovascular diseases: a ubiquitous dangerous molecule worthy of vigilance [J]. Front Cell Dev Biol, 2022, 9: 781839. |
| [53] | van der Werf MJ, Zeikus JG. 5-Aminolevulinate production by Escherichia coli containing the Rhodobacter sphaeroides hemA gene [J]. Appl Environ Microbiol, 1996, 62(10): 3560-3566. |
| [54] | Choi HP, Hong JW, Rhee KH, et al. Cloning, expression, and characterization of 5-aminolevulinic acid synthase from Rhodopseudomonas palustris KUGB306 [J]. FEMS Microbiol Lett, 2004, 236(2): 175-181. |
| [55] | Lou JW, Zhu L, Wu MB, et al. High-level soluble expression of the hemA gene from Rhodobacter capsulatus and comparative study of its enzymatic properties [J]. J Zhejiang Univ Sci B, 2014, 15(5): 491-499. |
| [56] | Wang WQ, Xiang YL, Yin GB, et al. Construction of 5-aminolevulinic acid microbial cell factories through identification of novel synthases and metabolic pathway screens and transporters [J]. J Agric Food Chem, 2024, 72(14): 8006-8017. |
| [57] | He GM, Jiang MR, Cui ZZ, et al. Construction of 5-aminolevulinic acid synthase variants by cysteine-targeted mutation to release heme inhibition [J]. J Biosci Bioeng, 2022, 134(5): 416-423. |
| [58] | Kim A, Stewart JD. Exploring the structure-function relationships in a 5-aminolevulinic acid synthase and the use of protein engineering to expand its substrate range [J]. Biochemistry, 2025, 64(1): 238-249. |
| [59] | Du S, Zheng N, Zhang ZH, et al. Rational design engineering of 5-aminolevulinate synthase with activity and stability enhancement [J]. J Agric Food Chem, 2025, 73(3): 1892-1901. |
| [60] | Yu TH, Yi YC, Shih IT, et al. Enhanced 5-aminolevulinic acid production by co-expression of codon-optimized hemA gene with chaperone in genetic engineered Escherichia coli [J]. Appl Biochem Biotechnol, 2020, 191(1): 299-312. |
| [61] | Miscevic D, Mao JY, Kefale T, et al. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli [J]. Biotechnol Bioeng, 2021, 118(1): 30-42. |
| [62] | Ren J, Zhou LB, Wang C, et al. An unnatural pathway for efficient 5-aminolevulinic acid biosynthesis with Glycine from glyoxylate based on retrobiosynthetic design [J]. ACS Synth Biol, 2018, 7(12): 2750-2757. |
| [63] | Zhu ZW, Fu B, Lu JJ, et al. Engineered production of 5-aminolevulinic acid in recombinant Escherichia coli BL21 [J]. Prep Biochem Biotechnol, 2025, 55(4): 446-456. |
| [64] | Cui ZY, Jiang ZN, Zhang JH, et al. Stable and efficient biosynthesis of 5-aminolevulinic acid using plasmid-free Escherichia coli [J]. J Agric Food Chem, 2019, 67(5): 1478-1483. |
| [65] | Ramzi AB, Hyeon JE, Kim SW, et al. 5-Aminolevulinic acid production in engineered Corynebacterium glutamicum via C5 biosynthesis pathway [J]. Enzyme Microb Technol, 2015, 81: 1-7. |
| [66] | Zhang B, Ye BC. Pathway engineering in Corynebacterium glutamicum S9114 for 5-aminolevulinic acid production [J]. 3 Biotech, 2018, 8(5): 247. |
| [67] | Man ZW, Xu MJ, Rao ZM, et al. Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production [J]. Sci Rep, 2016, 6: 28629. |
| [68] | Wen JB, Bao J. Engineering Corynebacterium glutamicum triggers glutamic acid accumulation in biotin-rich corn stover hydrolysate [J]. Biotechnol Biofuels, 2019, 12: 86. |
| [69] | Noh MH, Lim HG, Park S, et al. Precise flux redistribution to glyoxylate cycle for 5-aminolevulinic acid production in Escherichia coli [J]. Metab Eng, 2017, 43: 1-8. |
| [70] | Zhang JL, Weng HJ, Zhou ZX, et al. Engineering of multiple modular pathways for high-yield production of 5-aminolevulinic acid in Escherichia coli [J]. Bioresour Technol, 2019, 274: 353-360. |
| [71] | Yi Y-C, Xue CF, Ng IS. Low-carbon-footprint production of high-end 5-aminolevulinic acid via integrative strain engineering and RuBisCo-equipped Escherichia coli [J]. ACS Sustainable Chem Eng, 2021, 9(46): 15623-15633. |
| [72] | Lin H, San KY, Bennett GN. Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli [J]. Appl Microbiol Biotechnol, 2005, 67(4): 515-523. |
| [73] | Buch AD, Archana G, Kumar GN. Enhanced citric acid biosynthesis in Pseudomonas fluorescens ATCC 13525 by overexpression of the Escherichia coli citrate synthase gene [J]. Microbiology, 2009, 155(Pt 8): 2620-2629. |
| [74] | Lai PH, Ng IS. Accelerated 5-aminolevulinic acid biosynthesis by coupling aconitase and ALA synthase in engineered Escherichia coli [J]. Biochem Eng J, 2024, 209: 109419. |
| [75] | Effendi SSW, Ng IS. Non-native pathway engineering with CRISPRi for carbon dioxide assimilation and valued 5-aminolevulinic acid synthesis in Escherichia coli nissle [J]. ACS Synth Biol, 2024, 13(7): 2038-2044. |
| [76] | Ding WW, Weng HJ, Du GC, et al. 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli [J]. J Ind Microbiol Biotechnol, 2017, 44(8): 1127-1135. |
| [77] | Ge FL, Wen DM, Ren Y, et al. Downregulating of hemB via synthetic antisense RNAs for improving 5-aminolevulinic acid production in Escherichia coli [J]. 3 Biotech, 2021, 11(5): 230. |
| [78] | Yu XL, Jin HY, Liu WJ, et al. Engineering Corynebacterium glutamicum to produce 5-aminolevulinic acid from glucose [J]. Microb Cell Fact, 2015, 14: 183. |
| [79] | Su TY, Guo Q, Zheng Y, et al. Fine-tuning of hemB using CRISPRi for increasing 5-aminolevulinic acid production in Escherichia coli [J]. Front Microbiol, 2019, 10: 1731. |
| [80] | Zhang J, Wang ZG, Su TY, et al. Tuning the binding affinity of heme-responsive biosensor for precise and dynamic pathway regulation [J]. iScience, 2020, 23(5): 101067. |
| [81] | Yu XL, Jin HY, Cheng XL, et al. Transcriptomic analysis for elucidating the physiological effects of 5-aminolevulinic acid accumulation on Corynebacterium glutamicum [J]. Microbiol Res, 2016, 192: 292-299. |
| [82] | Elfsson B, Wallin I, Eksborg S, et al. Stability of 5-aminolevulinic acid in aqueous solution [J]. Eur J Pharm Sci, 1999, 7(2): 87-91. |
| [83] | Hunter GA, Rivera E, Ferreira GC. Supraphysiological concentrations of 5-aminolevulinic acid dimerize in solution to produce superoxide radical anions via a protonated dihydropyrazine intermediate [J]. Arch Biochem Biophys, 2005, 437(2): 128-137. |
| [84] | Bechara EJH, Dutra F, Cardoso VES, et al. The dual face of endogenous α-aminoketones: pro-oxidizing metabolic weapons [J]. Comp Biochem Physiol Part C Toxicol Pharmacol, 2007, 146(1-2): 88-110. |
| [85] | Wu J, Wu J, He RL, et al. Modularized engineering of Shewanella oneidensis MR-1 for efficient and directional synthesis of 5-aminolevulinic acid [J]. Metab Eng, 2024, 83: 206-215. |
| [86] | Pu W, Chen JZ, Zhou YY, et al. Systems metabolic engineering of Escherichia coli for hyper-production of 5-aminolevulinic acid [J]. Biotechnol Biofuels Bioprod, 2023, 16(1): 31. |
| [87] | Park SY, Yang D, Ha SH, et al. Metabolic engineering of microorganisms for the production of natural compounds [J]. Adv Biosys, 2018, 2(1): 1700190. |
| [88] | Xu P, Li LY, Zhang FM, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control [J]. Proc Natl Acad Sci USA, 2014, 111(31): 11299-11304. |
| [89] | Tan SI, You SC, Shih IT, et al. Quantification, regulation and production of 5-aminolevulinic acid by green fluorescent protein in recombinant Escherichia coli [J]. J Biosci Bioeng, 2020, 129(4): 387-394. |
| [90] | Su HF, Chen SJ, Chen XL, et al. Utilizing a high-throughput visualization screening technology to develop a genetically encoded biosensor for monitoring 5-aminolevulinic acid production in engineered Escherichia coli [J]. Biosens Bioelectron, 2025, 267: 116806. |
| [91] | 陈久洲, 王钰, 蒲伟, 等. 5-氨基乙酰丙酸生物合成技术的发展及展望 [J]. 合成生物学, 2021, 2(6): 1000-1016. |
| Chen JZ, Wang Y, Pu W, et al. Development and prospects of 5-aminolevulinic acid biosynthesis technology [J]. Synthetic Biology, 2021, 2(6): 1000-1016. |
| [1] | 刘语诗, 李镇, 邹宇琛, 汤维维, 李彬. 药用植物空间代谢组学研究进展[J]. 生物技术通报, 2025, 41(9): 22-31. |
| [2] | 李加仪, 李尽益, 白雪, 柏映国, 刘波, 张志伟. 稀有密码子串联介导的HemB表达弱化提升5-氨基乙酰丙酸的含量[J]. 生物技术通报, 2025, 41(8): 74-81. |
| [3] | 蔡如凤, 杨宇轩, 于基正, 李佳楠. 人工智能重塑蛋白质工程:从结构解析到合成生物学的算法革命[J]. 生物技术通报, 2025, 41(8): 1-10. |
| [4] | 高婧, 陈益存, 高暝, 赵耘霄, 汪阳东. 植物单宁合成调控及其对环境的响应机制[J]. 生物技术通报, 2025, 41(7): 49-59. |
| [5] | 吴娅, 姚润, 杨含婷, 刘微, 杨帅, 宋驰, 陈士林. 凤梨薄荷SDR基因家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(5): 175-185. |
| [6] | 鲁天怡, 李爱朋, 费强. 生物合成聚乳酸研究进展[J]. 生物技术通报, 2025, 41(4): 47-60. |
| [7] | 李晓明, 尚秀华, 王有霜, 吴志华. 植物中苯并噁嗪类化合物的研究进展[J]. 生物技术通报, 2025, 41(4): 9-20. |
| [8] | 张纪娇, 王会颖, 房欢, 张大伟. 维生素B12生物合成领域的十年回顾与技术进展[J]. 生物技术通报, 2025, 41(11): 62-74. |
| [9] | 何听雨, 逄雨, 张远洋, 孙雪, 李玉, 路福平, 李庆刚. 高产乳酰-N-三糖Ⅱ大肠杆菌菌株的构建[J]. 生物技术通报, 2025, 41(11): 143-152. |
| [10] | 周思延, 丁炜权, 董平, 翁含之, 胥睿睿, 康振. 大肠杆菌Nissle 1917的合成生物学平台开发及应用进展[J]. 生物技术通报, 2025, 41(11): 47-61. |
| [11] | 徐欣欣, 李彦君, 张伟, 黄火清, 罗会颖, 姚斌. 人造淀粉生物合成技术:进展、挑战与展望[J]. 生物技术通报, 2025, 41(11): 22-27. |
| [12] | 魏敏华, 李晓童, 姜亚文, 周飘飘, 汪凯, 孙浩, 芦楠, 张成林. |
| [13] | 杨熠辰, 朱宏宇, 苏小运, 王苑, 罗会颖, 田健, 姚斌, 黄火清, 张杰. 以果葡糖浆为底物高效合成肌醇细胞工厂的构建[J]. 生物技术通报, 2025, 41(11): 121-133. |
| [14] | 冀梦然, 张瑞英, 刘红丹, 冯伟萌, 刘秀玉, 马蕊, 陈随清. 转录组与代谢组联合分析南阳艾嫩叶与老叶的萜类成分差异[J]. 生物技术通报, 2025, 41(10): 277-291. |
| [15] | 张雨珊, 张雯雯, 刘岩, 申玉璞, 孙鲁, 黄伟红, 李中媛. 伏马毒素的污染现状、毒性作用机制及防控策略研究进展[J]. 生物技术通报, 2025, 41(10): 129-142. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||