








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 124-138.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0390
徐嘉妤1,2( ), 赵阁3, 唐叶2, 刘雯雯3, 彭清忠1(
), 赵阁3, 唐叶2, 刘雯雯3, 彭清忠1( ), 吴家和1,2(
), 吴家和1,2( )
)
收稿日期:2025-04-14
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
彭清忠,男,博士,教授,研究生导师,研究方向 :生物化学与分子生物学;E-mail: qzpengjsu@jsu.edu.cn作者简介:徐嘉妤,女,硕士研究生,研究方向 :生物化学与分子生物学;E-mail: 2022700533@stu.jsu.edu.cn
基金资助:
XU Jia-yu1,2( ), ZHAO Ge3, TANG Ye2, LIU Wen-wen3, PENG Qing-zhong1(
), ZHAO Ge3, TANG Ye2, LIU Wen-wen3, PENG Qing-zhong1( ), WU Jia-he1,2(
), WU Jia-he1,2( )
)
Received:2025-04-14
Published:2025-12-26
Online:2026-01-06
摘要:
目的 探讨陆地棉WRKY基因家族特征及WRKY转录因子在棉纤维生长发育中的功能,为阐明其分子机制提供依据。 方法 对GhWRKY基因家族进行全基因组鉴定,分析染色体定位、理化性质、亚细胞定位、基因结构和启动子元件,结合实时荧光定量分析GhWRKY基因家族在纤维发育过程中的表达模式,并利用病毒诱导基因沉默技术解析GhWRKY44的功能。 结果 在陆地棉全基因组中,共鉴定到225个WRKY基因家族成员,可进一步分为3个亚家族;该家族基因不均匀分布在26条染色体上。启动子区域含有大量植物激素、生长发育、胁迫响应和光响应相关顺式作用元件。8个GhWRKYs(GhWRKY11/14/31/48/133/149/ 160/224)在纤维不同发育时期存在优势表达;其中GhWRKY14和GhWRKY133这2个基因在纤维不同发育时期具有相似的表达模式和三维结构,为拟南芥WRKY44在陆地棉中的2个直系同源基因(拷贝),因此将其命名为GhWRKY44。而GhWRKY44基因的沉默能够显著抑制表皮毛的生长发育。 结论 系统分析了陆地棉WRKY基因家族,筛选出在纤维生长发育方面的关键基因GhWRKY44,为解析WRKY44在纤维生长发育方面的功能提供研究基础。
徐嘉妤, 赵阁, 唐叶, 刘雯雯, 彭清忠, 吴家和. 陆地棉WRKY基因家族全基因组鉴定及WRKY44在纤维发育中的表达分析[J]. 生物技术通报, 2025, 41(12): 124-138.
XU Jia-yu, ZHAO Ge, TANG Ye, LIU Wen-wen, PENG Qing-zhong, WU Jia-he. Genome-wide Identification of WRKY Gene Family in Gossypium hirsutum and Expression Analysis of WRKY44 in Fiber Development[J]. Biotechnology Bulletin, 2025, 41(12): 124-138.
| 引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| qGhhis3-F | GACACCAACCTTTGCGCGAT |
| qGhhis3-R | AGCGACTGATCCACACTTCTG |
| qGhWRKY11-F | CCGAGCTCTTGGACTCTCCTGT |
| qGhWRKY11-R | TGTTGCGCAGTTTGATTTGCGT |
| qGhWRKY14-F | AGGCAGAGCAGTCCGGAATAGG |
| qGhWRKY14-R | TGATACAAGCTTTGCCTGCGGT |
| qGhWRKY31-F | GCTCGATTACCCCACGCTTCAA |
| qGhWRKY31-R | GCCCAGTAGGCGCAGTGAATAA |
| qGhWRKY48-F | GTCTCCTCCTGTAGCAAGCCAG |
| qGhWRKY48-R | ACACAGGTGTCTTCACAACGGG |
| qGhWRKY133-F | GGTTGTCTGTGCTTCCGGCTTA |
| qGhWRKY133-R | GCTCCCTCCTTGCTTGCTTCTT |
| qGhWRKY149-F | CTTCTGCTGGGTGTCCTGTACG |
| qGhWRKY149-R | GAGCAATTGGTTGGCCATGTCG |
| qGhWRKY160-F | TGGTCAGAAGCAAGTGAAGAGTCC |
| qGhWRKY160-R | TCTCAATTACATGGCCCGTCTGA |
| qWRKY224-F | GATGGGTATCGATGGCGCAAGT |
| qWRKY224-R | TGCTTTCGGATCATGAGACGCC |
表1 引物名称和序列
Table 1 Primer names and sequences
| 引物名称 Primer name | 序列 Sequence (5′‒3′) |
|---|---|
| qGhhis3-F | GACACCAACCTTTGCGCGAT |
| qGhhis3-R | AGCGACTGATCCACACTTCTG |
| qGhWRKY11-F | CCGAGCTCTTGGACTCTCCTGT |
| qGhWRKY11-R | TGTTGCGCAGTTTGATTTGCGT |
| qGhWRKY14-F | AGGCAGAGCAGTCCGGAATAGG |
| qGhWRKY14-R | TGATACAAGCTTTGCCTGCGGT |
| qGhWRKY31-F | GCTCGATTACCCCACGCTTCAA |
| qGhWRKY31-R | GCCCAGTAGGCGCAGTGAATAA |
| qGhWRKY48-F | GTCTCCTCCTGTAGCAAGCCAG |
| qGhWRKY48-R | ACACAGGTGTCTTCACAACGGG |
| qGhWRKY133-F | GGTTGTCTGTGCTTCCGGCTTA |
| qGhWRKY133-R | GCTCCCTCCTTGCTTGCTTCTT |
| qGhWRKY149-F | CTTCTGCTGGGTGTCCTGTACG |
| qGhWRKY149-R | GAGCAATTGGTTGGCCATGTCG |
| qGhWRKY160-F | TGGTCAGAAGCAAGTGAAGAGTCC |
| qGhWRKY160-R | TCTCAATTACATGGCCCGTCTGA |
| qWRKY224-F | GATGGGTATCGATGGCGCAAGT |
| qWRKY224-R | TGCTTTCGGATCATGAGACGCC |
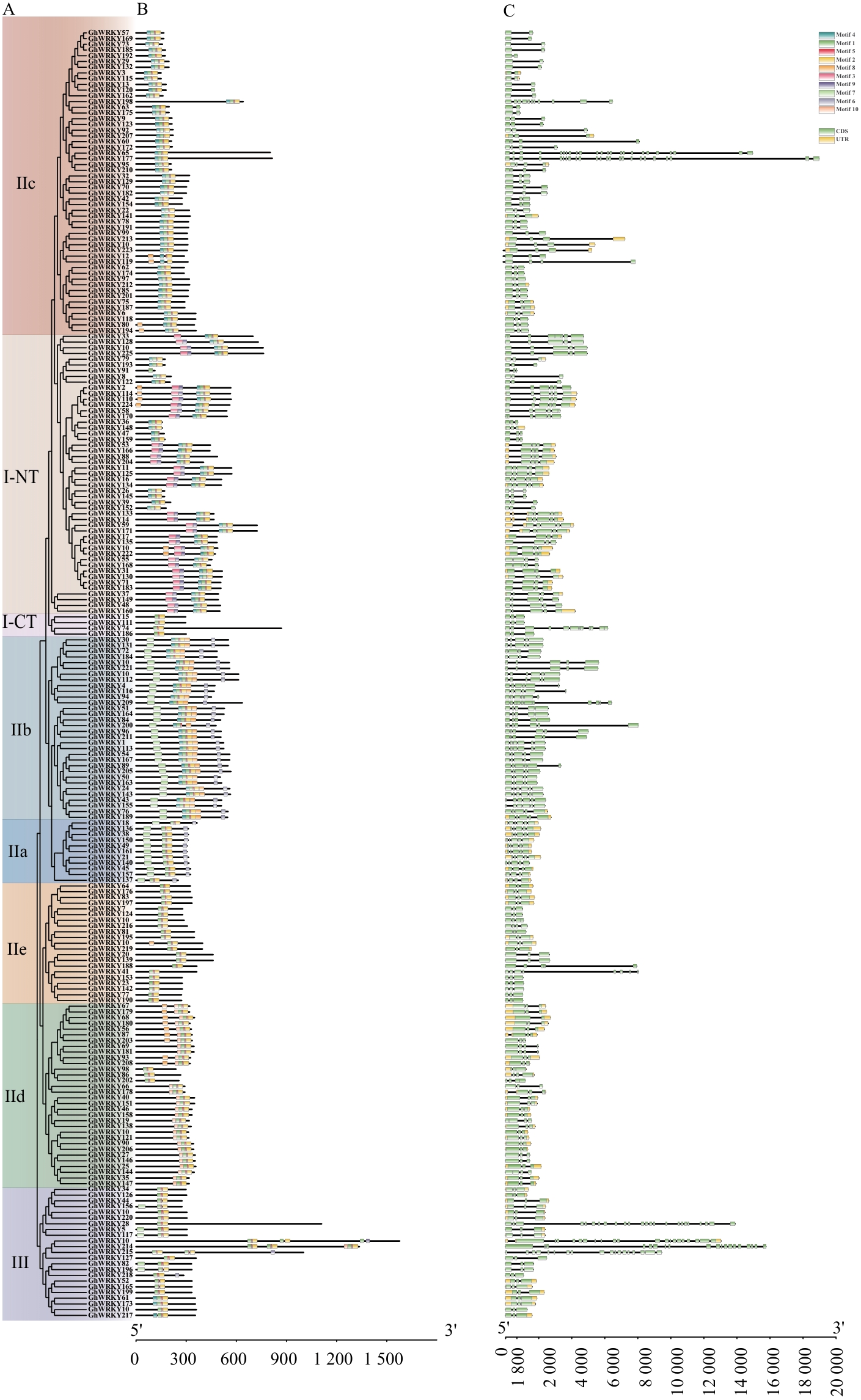
图4 GhWRKY基因家族成员保守基序和基因结构分析A:陆地棉GhWRKYs家族蛋白进化树;B:陆地棉GhWRKYs家族蛋白的保守基序;C:陆地棉GhWRKY基因家族的基因结构
Fig. 4 Conserved motifs and gene structure analysis of GhWRKY gene family membersA: Phylogenetic tree of the GhWRKY family protein in G. hirsutum. B: Conserved motif of GhWRKY proteins in G. hirsutum. C: Gene structure of the GhWRKY gene family in G. hirsutum
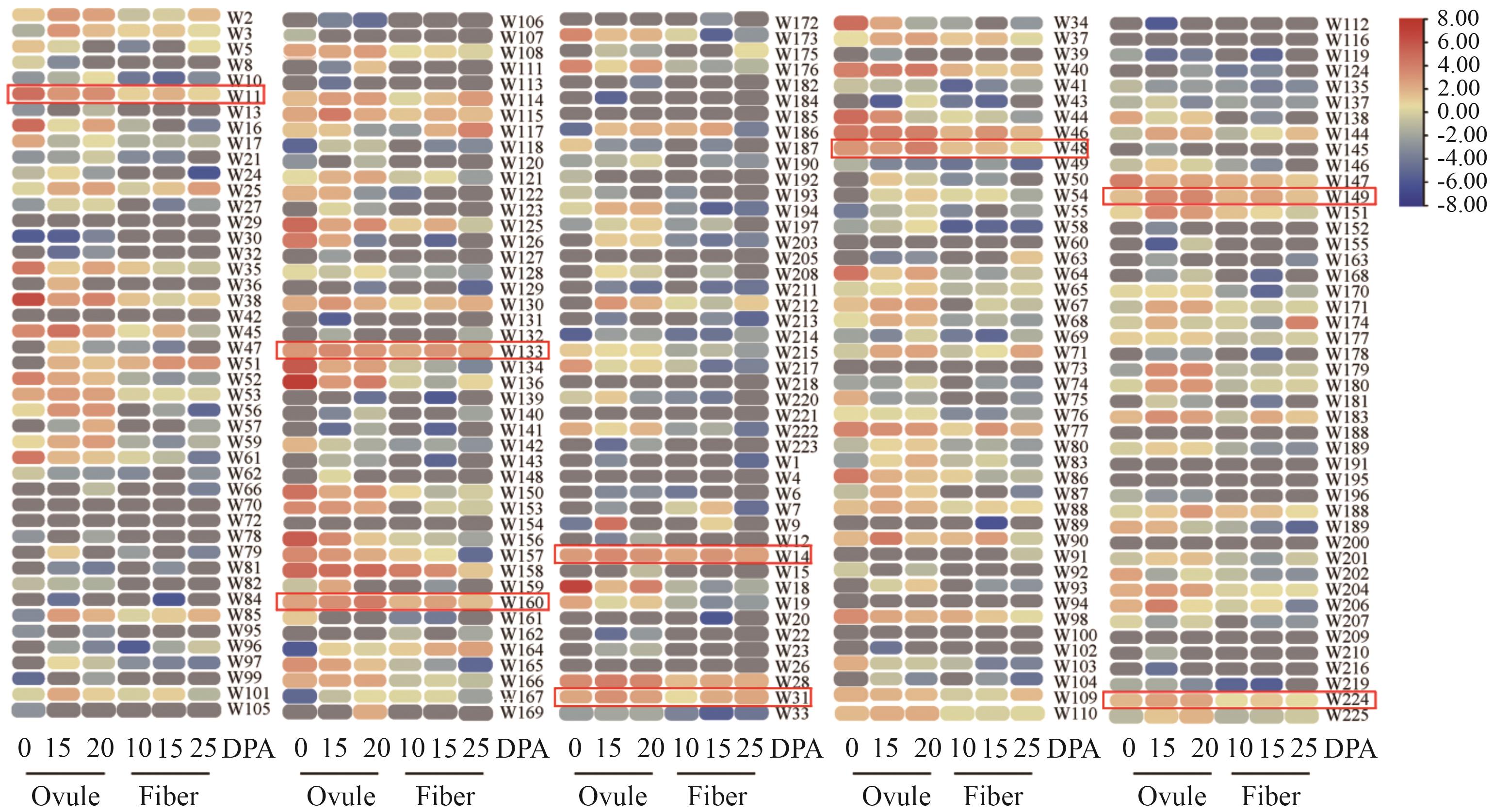
图6 GhWRKYs在纤维发育时期的表达量分析红框中标注的为纤维和胚珠不同发育时期优势表达基因。DPA:开花后天数。下同
Fig. 6 Expression analysis of GhWRKYs during fiber developmentThe genes highlighted in red boxes are predominantly expressed during different developmental stages of fiber and ovule. DPA: Day post anthesis. The same below
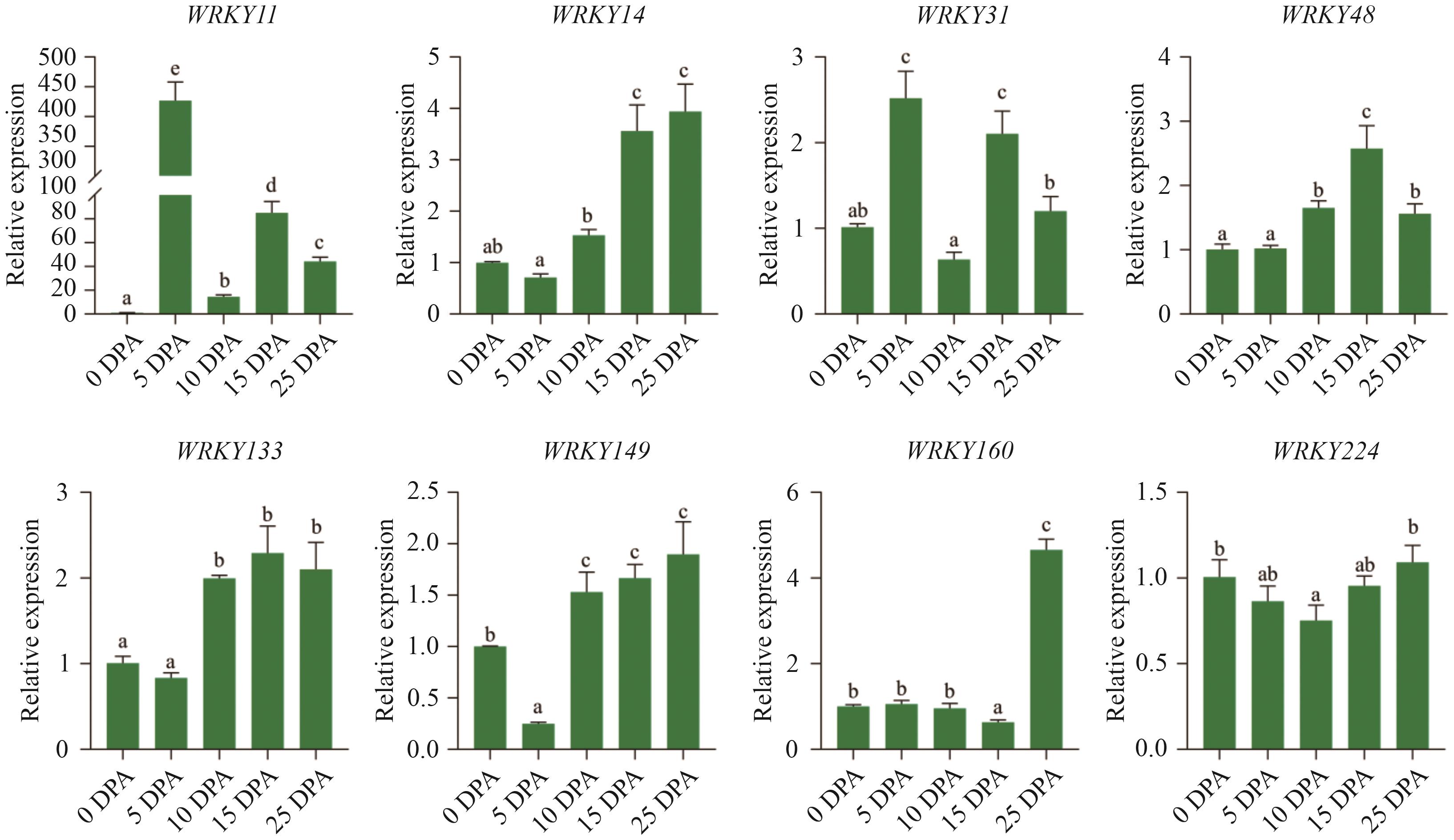
图7 GhWRKYs在棉纤维发育不同时期的表达量分析采用Tukey test进行统计分析,不同小写字母表示GhWRKYs在棉纤维发育各阶段表达量差异显著(P<0.05)
Fig. 7 Relative expressions of key GhWRKYs during different stages of cotton fiber developmentStatistical analyses were performed using the Tukey test. Different lower letters indicate significant difference of the expression of GhWRKYs at different stages of cotton fiber development (P<0.05)
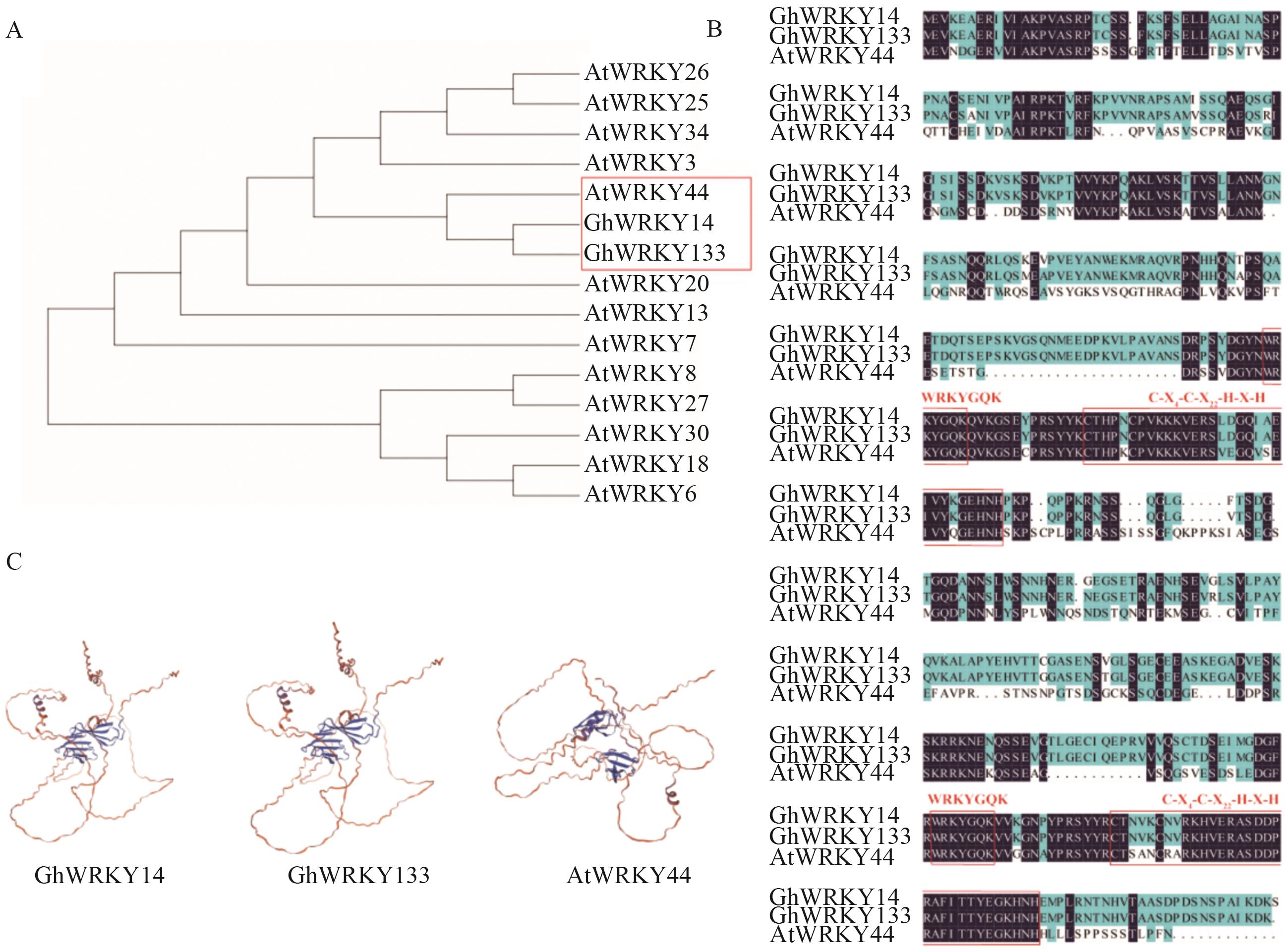
图8 GhWRKY44同源分析A:GhWRKY14(Gh_A04G096000.1)、GhWRKY133(Gh_D04G134100.1)和AtWRKY44(AT2G37260)系统进化树;B:GhWRKY14、GhWRKY133和AtWRKY44氨基酸序列比对分析;C:GhWRKY14、GhWRKY133和AtWRKY44蛋白三维结构
Fig. 8 Homology analysis of GhWRKY44A: Phylogenetic tree of GhWRKY14 (Gh_A04G096000.1), GhWRKY133 (Gh_D04G134100.1) and AtWRKY44 (AT2G37260). B: Amino acid sequence comparison analysis of GhWRKY14, GhWRKY133 and AtWRKY44. C: 3D structures of GhWRKY14, GhWRKY133 and AtWRKY44 proteins
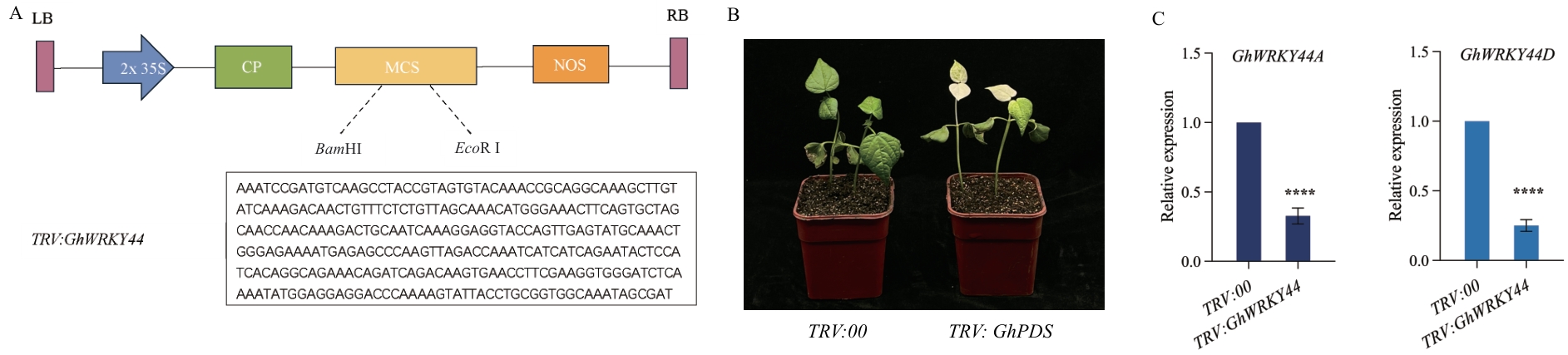
图10 TRV:GhWRKY44沉默植株的培育A:VIGS沉默载体示意图;B:农杆菌侵染棉花14 d后,TRV:GhPDS植株表现出明显的白化表型;C:RT-qPCR检测GhWRKY44沉默效率,n=35;以GhHis3为内参基因,误差线为3次生物学重复的标准误;采用独立方差t检验进行统计分析,*P<0.05,**P<0.01,***P<0.001,****P<0.0001。下同
Fig. 10 Cultivating of TRV:GhWRKY44-silenced plantsA: Schematic diagram of VIGS silencing vector. B: Fourteen days after Agrobacterium infestation of cotton, TRV:GhPDS plants showed a distinct albino phenotype. C: RT-qPCR detection of GhWRKY44, silencing efficiency, n=35. GhHis3 was used as the internal reference gene, and the error line was the standard error of three biological replicates. Statistical analysis was performed using an independent variance t-test, * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. The same below
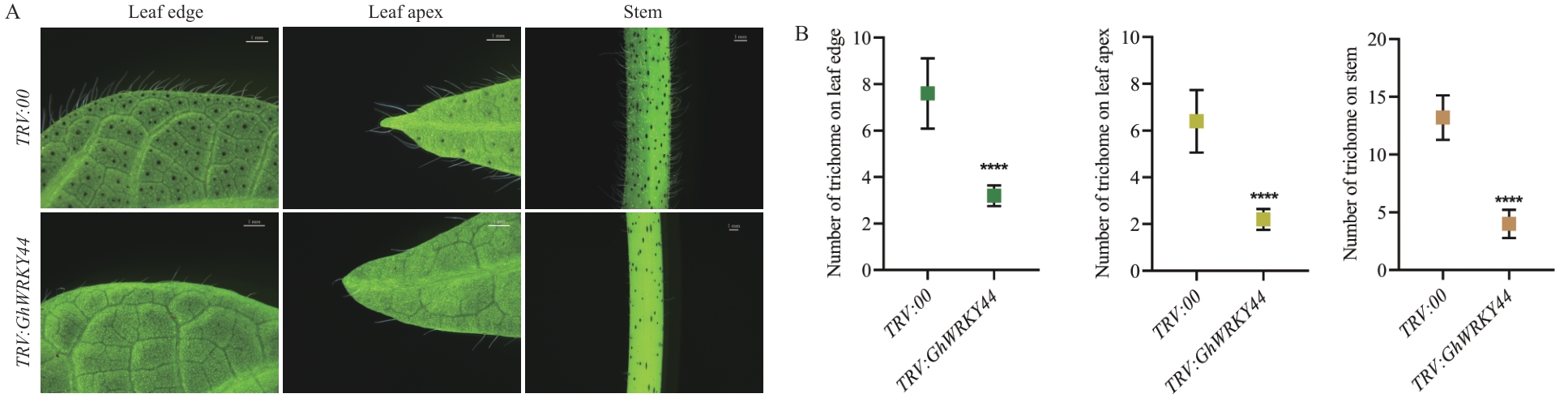
图11 GhWRKY44的沉默株系中表皮毛的表型鉴定A:GhWRKY44沉默植株的表皮毛形态;B:GhWRKY44沉默植株的表皮毛密度统计分析,n=15
Fig. 11 Phenotypic characterization of epidermal trichomes in GhWRKY44-silenced plantsA: Trichome morphology of GhWRKY44-silenced plants. B: Statistical analysis of trichome density of GhWRKY44-silenced plants, n=15
| [1] | Qin Y, Sun ML, Li WW, et al. Single-cell RNA-seq reveals fate determination control of an individual fibre cell initiation in cotton (Gossypium hirsutum) [J]. Plant Biotechnol J, 2022, 20(12): 2372-2388. |
| [2] | Tian HD, Wang QL, Yan XY, et al. The disruptions of sphingolipid and sterol metabolism in the short fiber of Ligon-lintless-1 mutant revealed obesity impeded cotton fiber elongation and secondary cell wall deposition [J]. Int J Mol Sci, 2025, 26(3): 1375. |
| [3] | Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis [J]. Plant Physiol, 2001, 127(4): 1361-1366. |
| [4] | Haigler CH, Betancur L, Stiff MR, et al. Cotton fiber: a powerful single-cell model for cell wall and cellulose research [J]. Front Plant Sci, 2012, 3: 104. |
| [5] | Wu N, Yang J, Wang GN, et al. Novel insights into water-deficit-responsive mRNAs and lncRNAs during fiber development in Gossypium hirsutum [J]. BMC Plant Biol, 2022, 22(1): 6. |
| [6] | Sun XW, Qin AZ, Wang XX, et al. Spatiotemporal transcriptome and metabolome landscapes of cotton fiber during initiation and early development [J]. Nat Commun, 2025, 16(1): 858. |
| [7] | Wani SH, Anand S, Singh B, et al. WRKY transcription factors and plant defense responses: latest discoveries and future prospects [J]. Plant Cell Rep, 2021, 40(7): 1071-1085. |
| [8] | Song H, Cao YP, Zhao LG, et al. Review: WRKY transcription factors: Understanding the functional divergence [J]. Plant Sci, 2023, 334: 111770. |
| [9] | Eulgem T, Rushton PJ, Robatzek S, et al. The WRKY superfamily of plant transcription factors [J]. Trends Plant Sci, 2000, 5(5): 199-206. |
| [10] | Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1 that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato [J]. Mol Gen Genet, 1994, 244(6): 563-571. |
| [11] | Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function [J]. Curr Opin Plant Biol, 2004, 7(5): 491-498. |
| [12] | Sahebi M, Hanafi MM, Rafii MY, et al. Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family [J]. Biomed Res Int, 2018, 2018: 3158474. |
| [13] | Hu WJ, Ren QY, Chen YL, et al. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses [J]. BMC Plant Biol, 2021, 21(1): 427. |
| [14] | Xing LN, Zhang YF, Ge MR, et al. Identification of WRKY gene family in Dioscorea opposita Thunb. reveals that DoWRKY71 enhanced the tolerance to cold and ABA stress [J]. Peer J, 2024, 12: e17016. |
| [15] | Lei L, Dong K, Liu SW, et al. Genome-wide identification of the WRKY gene family in blueberry (Vaccinium spp.) and expression analysis under abiotic stress [J]. Front Plant Sci, 2024, 15: 1447749. |
| [16] | Xiong R, Peng Z, Zhou H, et al. Correction: Genome-wide identification, structural characterization and gene expression analysis of the WRKY transcription factor family in pea (Pisum sativum L.) [J]. BMC Plant Biol, 2024, 24(1): 141. |
| [17] | Zhao H, Wang S, Chen S, et al. Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii × Populus nigra [J]. Gene, 2015, 565(1): 130-139. |
| [18] | Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2 a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor [J]. Plant Cell, 2002, 14(6): 1359-1375. |
| [19] | Liu R, Xu YH, Jiang SC, et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function [J]. J Exp Bot, 2013, 64(18): 5443-5456. |
| [20] | Ding ZJ, Yan JY, Li GX, et al. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA [J]. Plant J, 2014, 79(5): 810-823. |
| [21] | Lei RH, Ma ZB, Yu DQ. WRKY2/34-VQ20 modules in Arabidopsis thaliana negatively regulate expression of a trio of related MYB transcription factors during pollen development [J]. Front Plant Sci, 2018, 9: 331. |
| [22] | Li W, Wang HP, Yu DQ. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions [J]. Mol Plant, 2016, 9(11): 1492-1503. |
| [23] | Ma ZB, Li W, Wang HP, et al. WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering [J]. J Integr Plant Biol, 2020, 62(11): 1659-1673. |
| [24] | Zhang LP, Chen LG, Yu DQ. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering [J]. Plant Physiol, 2018, 176(1): 790-803. |
| [25] | Rishmawi L, Pesch M, Juengst C, et al. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis [J]. Plant Physiol, 2014, 165(1): 186-195. |
| [26] | Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense [J]. Genes Dev, 2002, 16(9): 1139-1149. |
| [27] | Doll J, Muth M, Riester L, et al. Arabidopsis thaliana WRKY25 transcription factor mediates oxidative stress tolerance and regulates senescence in a redox-dependent manner [J]. Front Plant Sci, 2020, 10: 1734. |
| [28] | Chen LG, Xiang SY, Chen YL, et al. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence [J]. Mol Plant, 2017, 10(9): 1174-1189. |
| [29] | Jiang YJ, Yu DQ. The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise Botrytis cinerea resistance [J]. Plant Physiol, 2016, 171(4): 2771-2782. |
| [30] | Wu XL, Shiroto Y, Kishitani S, et al. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter [J]. Plant Cell Rep, 2009, 28(1): 21-30. |
| [31] | He GH, Xu JY, Wang YX, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis [J]. BMC Plant Biol, 2016, 16(1): 116. |
| [32] | Li FJ, Li MY, Wang P, et al. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59 [J]. New Phytol, 2017, 215(4): 1462-1475. |
| [33] | Li SJ, Fu QT, Chen LG, et al. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance [J]. Planta, 2011, 233(6): 1237-1252. |
| [34] | Zhang Y, Yu HJ, Yang XY, et al. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner [J]. Plant Physiol Biochem, 2016, 108: 478-487. |
| [35] | Yokotani N, Sato Y, Tanabe S, et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance [J]. J Exp Bot, 2013, 64(16): 5085-5097. |
| [36] | Wang NN, Li Y, Chen YH, et al. Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum) [J]. Plant Cell, 2021, 33(8): 2736-2752. |
| [37] | Yang L, Qin WQ, Wei X, et al. Regulatory networks of coresident subgenomes during rapid fiber cell elongation in upland cotton [J]. Plant Commun, 2024, 5(12): 101130. |
| [38] | Yang DJ, Liu YY, Cheng HL, et al. Identification of the group ⅢWRKY subfamily and the functional analysis of GhWRKY53 in Gossypium hirsutum L [J]. Plants, 2021, 10(6): 1235. |
| [39] | Wang HP, Chen WQ, Xu ZY, et al. Functions of WRKYs in plant growth and development [J]. Trends Plant Sci, 2023, 28(6): 630-645. |
| [40] | Sun PW, Xu YH, Yu CC, et al. WRKY44 represses expression of the wound-induced sesquiterpene biosynthetic gene ASS1 in Aquilaria sinensis [J]. J Exp Bot, 2020, 71(3): 1128-1138. |
| [41] | Peng YY, Thrimawithana AH, Cooney JM, et al. The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway [J]. Sci Rep, 2020, 10(1): 14161. |
| [42] | Han Y, Zhang X, Wang W, et al. Correction: the suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana [J]. PLoS One, 2015, 10(4): e0124854. |
| [43] | Li QY, Yin M, Li YP, et al. Expression of Brassica napus TTG2 a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes [J]. J Exp Bot, 2015, 66(19): 5821-5836. |
| [44] | Wen TY, Xu X, Ren AP, et al. Genome-wide identification of terpenoid synthase family genes in Gossypium hirsutum and functional dissection of its subfamily cadinene synthase A in gossypol synthesis [J]. Front Plant Sci, 2023, 14: 1162237. |
| [45] | Bencke-Malato M, Cabreira C, Wiebke-Strohm B, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection [J]. BMC Plant Biol, 2014, 14: 236. |
| [46] | Shi GF, Liu GL, Liu HJ, et al. WRKY transcriptional factor IlWRKY70 from Iris laevigata enhances drought and salinity tolerances in Nicotiana tabacum [J]. Int J Mol Sci, 2023, 24(22): 16174. |
| [47] | Zhang JW, Huang DZ, Zhao XJ, et al. Drought-responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica enhance drought resistance in transgenic Arabidopsis [J]. Front Plant Sci, 2022, 13: 983600. |
| [48] | Chen JN, Nolan TM, Ye HX, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses [J]. Plant Cell, 2017, 29(6): 1425-1439. |
| [49] | Shim JS, Jung C, Lee S, et al. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling [J]. Plant J, 2013, 73(3): 483-495. |
| [50] | Turck F, Zhou AF, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in Parsley [J]. Plant Cell, 2004, 16(10): 2573-2585. |
| [1] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [2] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [3] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [4] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [5] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [6] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [7] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [8] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [9] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [10] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [11] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [12] | 张勇, 宋盛龙, 李永泰, 张新宇, 李艳军. 陆地棉GhSWEET9基因的克隆及抗黄萎病功能分析[J]. 生物技术通报, 2025, 41(6): 144-154. |
| [13] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [14] | 瞿美玲, 周思敏, 张惊宇, 何佳蔚, 朱佳源, 刘笑蓉, 童巧珍, 周日宝, 刘湘丹. 灰毡毛忍冬bHLH转录基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(6): 256-268. |
| [15] | 刘鑫, 王嘉雯, 李进伟, 牟策, 杨盼盼, 明军, 徐雷锋. 兰州百合三个LdBBXs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(5): 186-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||