








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 328-341.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0491
赵逸凡1( ), 王彤1(
), 王彤1( ), 叶岚2, 赵乐1, 张宝1, 杜鹏强3(
), 叶岚2, 赵乐1, 张宝1, 杜鹏强3( ), 何海荣1(
), 何海荣1( )
)
收稿日期:2025-05-13
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
何海荣,博士,副教授,研究方向 :微生物菌种鉴定;E-mail: hhirong@163.com作者简介:赵逸凡,女,研究方向 :微生物菌种鉴定;E-mail: xi040122@163.com基金资助:
ZHAO Yi-fan1( ), WANG Tong1(
), WANG Tong1( ), YE Lan2, ZHAO Le1, ZHANG Bao1, DU Peng-qiang3(
), YE Lan2, ZHAO Le1, ZHANG Bao1, DU Peng-qiang3( ), HE Hai-rong1(
), HE Hai-rong1( )
)
Received:2025-05-13
Published:2025-12-26
Online:2026-01-06
摘要:
目的 分离筛选抗地黄根腐病的生防菌株,为地黄根腐病的生物防治提供菌株资源。 方法 从感染轮纹病的地黄根际土中分离纯化微生物,采用平板对峙法筛选拮抗菌株,并通过盆栽试验进一步明确菌株对地黄根腐病的防治效果;结合形态观察、生理生化及分子生物学等方法确定菌株的分类地位;并对该菌株开展全基因组测序,分析基因组信息,探究其抗病机制。 结果 根据平板对峙结果,筛选到一株对植物病原真菌有广谱拮抗活性的菌株QH-1,其对地黄根腐病原菌的抑菌率接近90%。经鉴定,QH-1为一株多粘类芽胞杆菌(Paenibacillus polymyxa),为革兰氏阳性菌,产花生状芽孢。其在10-55 ℃,pH 6-8,NaCl浓度为0-5%的条件下均能生长。碳氮源利用实验表明,其可以利用多种氮源,而仅能利用麦芽糖和山梨醇两种碳源。另外,该菌株能产生过氧化氢酶、淀粉酶、酯酶、明胶酶、凝乳酶和蛋白酶。QH-1菌株的基因组信息表明其由一条环状染色体(5 692 874 bp)和一个环状质粒(37 590 bp)组成,(G+C)%含量为45.4%;antiSMASH结果显示基因组中含有17个生物合成基因簇,其中6个生物合成基因簇与6种抗菌化合物(fusaricidin B、paenibacillin、paenilan、tridecaptin、polymyxin和paenicidin A)的生物合成基因簇的相似性为100%,说明菌株QH-1具有产生以上抗菌活性化合物的潜力。 结论 菌株QH-1是一株抗地黄根腐病的多粘类芽胞杆菌,其基因组信息显示其能产生多种抗菌物质,为进一步研发抗地黄根腐病的微生物菌剂和天然抗菌物质提供了良好的理论和物质基础。
赵逸凡, 王彤, 叶岚, 赵乐, 张宝, 杜鹏强, 何海荣. 地黄根腐病拮抗类芽胞杆菌的筛选、鉴定和全基因组分析[J]. 生物技术通报, 2025, 41(12): 328-341.
ZHAO Yi-fan, WANG Tong, YE Lan, ZHAO Le, ZHANG Bao, DU Peng-qiang, HE Hai-rong. Screening, Identification and Whole Genome Analysis of a Paenibacillus Strain Resistant to Root Rot of Rehmannia glutinosa[J]. Biotechnology Bulletin, 2025, 41(12): 328-341.
病原真菌 Pathogenic fungi | 抑菌率 Fungal inhibition rate (%) | ||||||
|---|---|---|---|---|---|---|---|
| QH-1 | QH-9 | QH-16 | QH-33 | QH-39 | QH-41 | ||
| 褐斑病菌 C. cassiicola | 59.07±5.45cd | 45.73±0.84b | 40.13 1.75e | 35.7±2.65d | 49.63±3.65c | NA | |
| 茎基腐病菌 F. graminearum Schw | 88.84±3.49b | NA | 35.76±2.78d | 38.65±2.07c | NA | 47.55±3.98b | |
| 炭疽病菌 C. orbiculare | 51.44±3.50d | 65.51±3.03a | NA | NA | 49.78±4.97d | 40.20±3.69c | |
| 枯萎病菌 F. oxysporum | 15.47±5.11e | NA | 20.36±1.06c | NA | NA | NA | |
| 叶斑病菌 F. solani | 80.28±5.00c | NA | NA | NA | NA | 55.65±6.01d | |
| 黄萎病菌 V. dahliae Kleb | 51.80±3.78d | NA | 42.76±6.7b | 38.95±4.88b | NA | NA | |
| 轮纹病菌 P. herbarum | 89.27±5.25c | 34.77±4.32c | NA | 58.76±5.68a | 42.65±2.09a | NA | |
| 根腐病菌 Fusarium sp. Rf7 | 89.57±2.97a | NA | 60.23±3.54a | NA | 75.23±6.35a | 65.30±5.07a | |
表1 6株菌对植物病原真菌的抑菌率
Table 1 Antifungal activity of 6 bacterial strains against plant pathogenic fungi
病原真菌 Pathogenic fungi | 抑菌率 Fungal inhibition rate (%) | ||||||
|---|---|---|---|---|---|---|---|
| QH-1 | QH-9 | QH-16 | QH-33 | QH-39 | QH-41 | ||
| 褐斑病菌 C. cassiicola | 59.07±5.45cd | 45.73±0.84b | 40.13 1.75e | 35.7±2.65d | 49.63±3.65c | NA | |
| 茎基腐病菌 F. graminearum Schw | 88.84±3.49b | NA | 35.76±2.78d | 38.65±2.07c | NA | 47.55±3.98b | |
| 炭疽病菌 C. orbiculare | 51.44±3.50d | 65.51±3.03a | NA | NA | 49.78±4.97d | 40.20±3.69c | |
| 枯萎病菌 F. oxysporum | 15.47±5.11e | NA | 20.36±1.06c | NA | NA | NA | |
| 叶斑病菌 F. solani | 80.28±5.00c | NA | NA | NA | NA | 55.65±6.01d | |
| 黄萎病菌 V. dahliae Kleb | 51.80±3.78d | NA | 42.76±6.7b | 38.95±4.88b | NA | NA | |
| 轮纹病菌 P. herbarum | 89.27±5.25c | 34.77±4.32c | NA | 58.76±5.68a | 42.65±2.09a | NA | |
| 根腐病菌 Fusarium sp. Rf7 | 89.57±2.97a | NA | 60.23±3.54a | NA | 75.23±6.35a | 65.30±5.07a | |
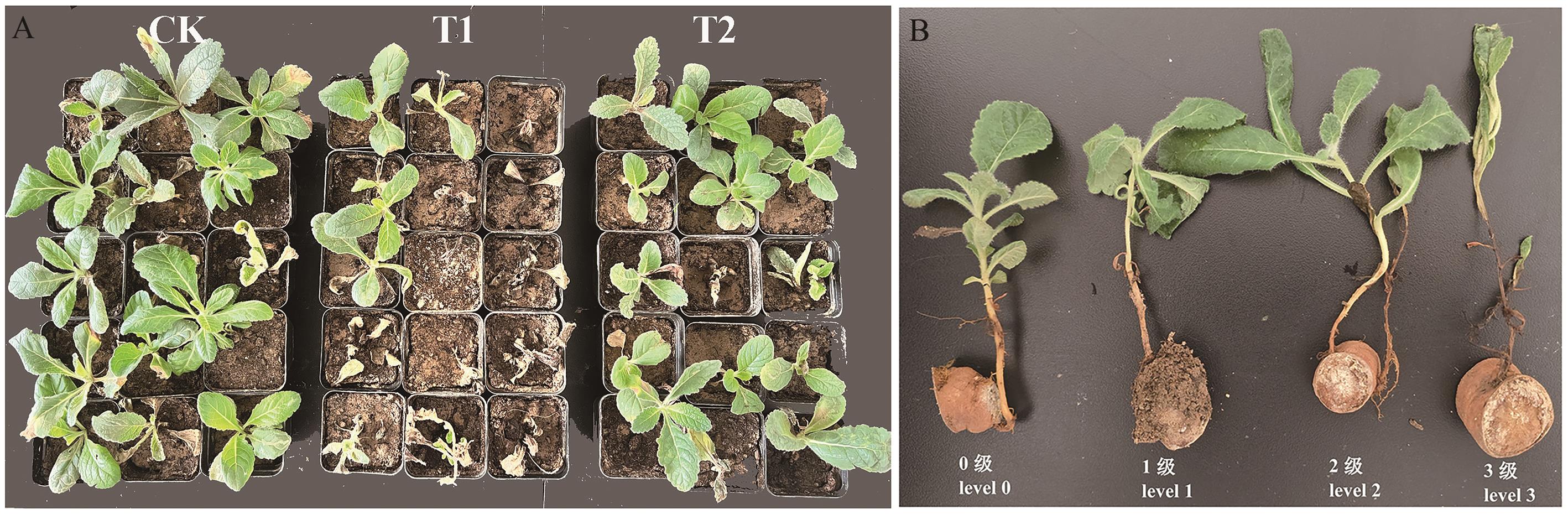
图2 QH-1对地黄根腐病防效实验中地黄盆栽情况(A)及地黄根腐病病害级数(B)
Fig. 2 Pot cultivation conditions of R. glutinosa in the QH-1 against R. glutinosa root rot (A) and disease severity level of R. glutinosa root rot (B)
| 处理 Treatment | 病情指数 Disease index | 发病率 Incidence (%) | 防病效果 Prevention effect (%) |
|---|---|---|---|
| CK(无菌水处理) Treatment with sterile water | 10.12 ± 3.16c | 15.56 ± 4.71c | 46.82 ± 11.75 |
| T1(Rf7处理) Treatment with pathogen Rf7 | 54.32 ± 4.45a | 74.07 ± 9.69a | |
| T2(Rf7+QH-1处理) Treatment with Rf7 and QH-1 | 28.64 ± 5.82b | 49.63 ± 5.88b |
表2 QH-1对地黄根腐病的防治效果
Table 2 Control effect of QH-1 against Fusarium root rot of R. glutinosa
| 处理 Treatment | 病情指数 Disease index | 发病率 Incidence (%) | 防病效果 Prevention effect (%) |
|---|---|---|---|
| CK(无菌水处理) Treatment with sterile water | 10.12 ± 3.16c | 15.56 ± 4.71c | 46.82 ± 11.75 |
| T1(Rf7处理) Treatment with pathogen Rf7 | 54.32 ± 4.45a | 74.07 ± 9.69a | |
| T2(Rf7+QH-1处理) Treatment with Rf7 and QH-1 | 28.64 ± 5.82b | 49.63 ± 5.88b |
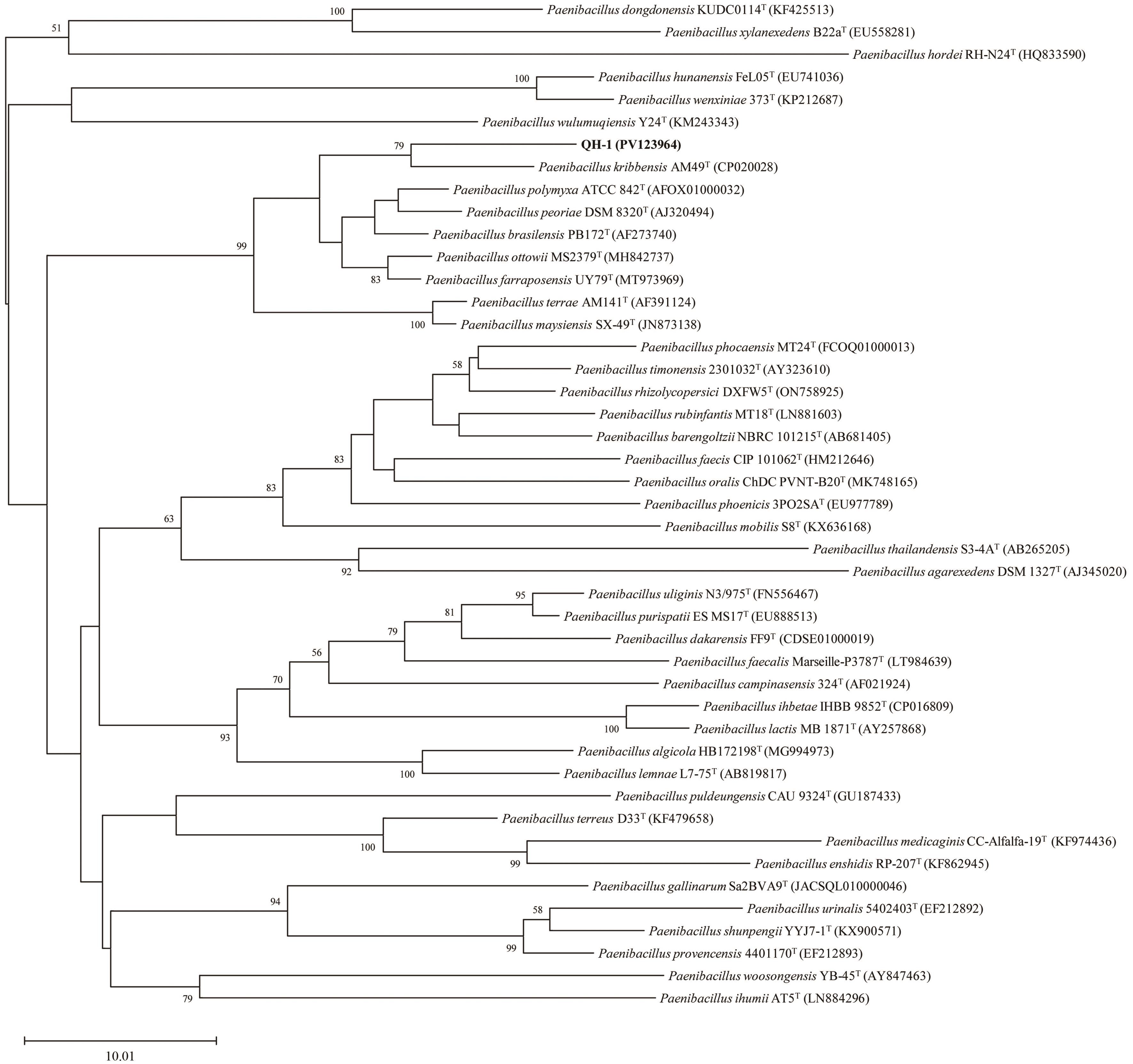
图3 菌株QH-1基于16S rRNA基因序列构建的邻接树分支节点处显示基于1 000次重复抽样、自举值>50%的支持率。标尺:每个核苷酸位点10.01个替换
Fig. 3 Neighbour-joining tree showing the phylogenetic position of strain QH-1 based on 16S rRNA gene sequencesBootstrap values >50% (based on 1 000 replications) are shown at branch points. Bar: 10.01 substitutions per nucleotide position
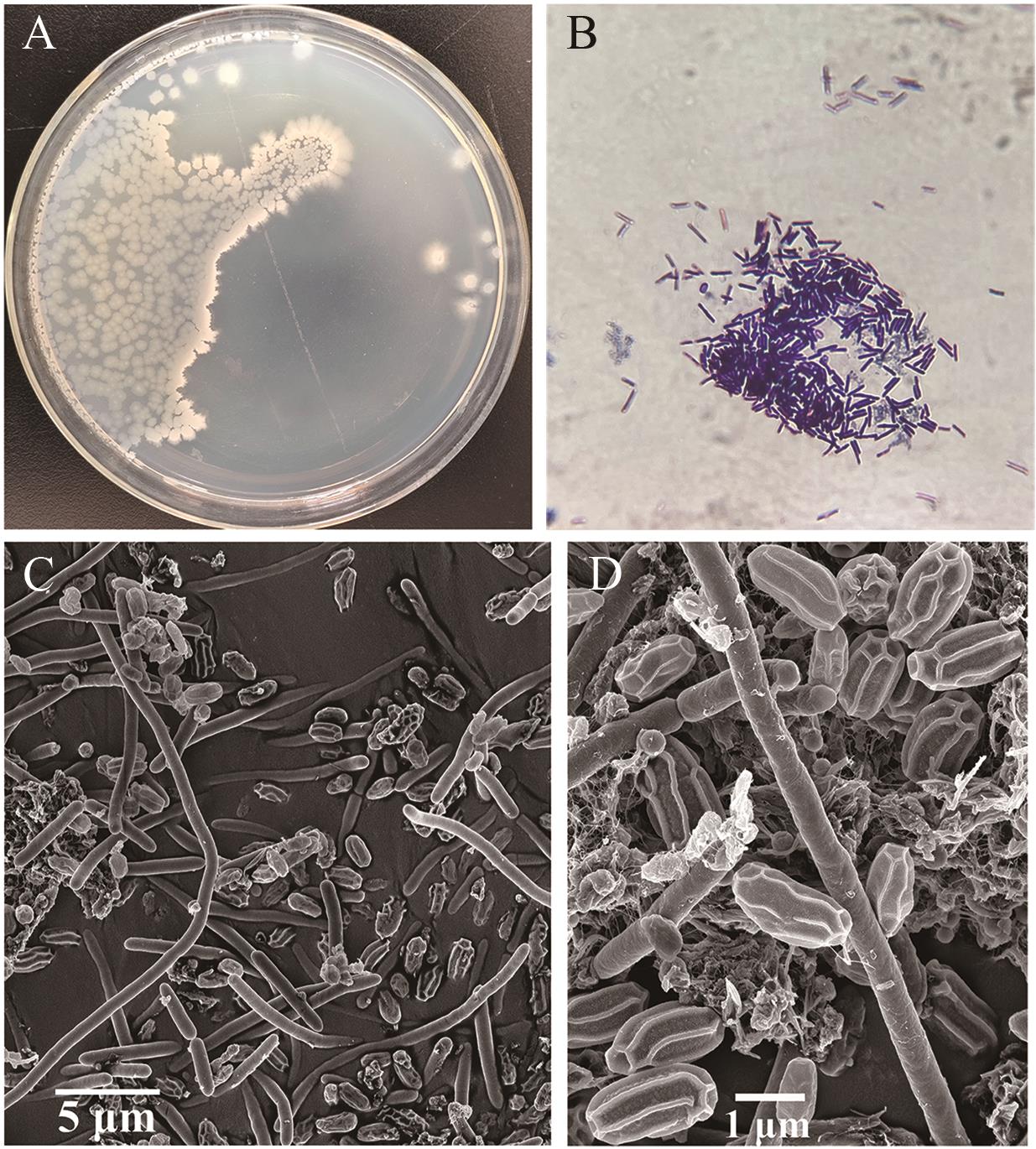
图4 QH-1的形态A: 平板上的形态; B: 100倍下QH-1菌株的革兰氏染色; C: 2 000倍扫描电镜图,bar 5 μm; D: 10 000倍扫描电镜,bar 1 μm
Fig. 4 Morphology of QH-1A: Agar plate; B: Gram stain of QH-1 in 100×; C: SEM image at 2 000×, bar 5 μm; D: SEM image at 10 000×, bar 1 μm
| 特征Characteristic | QH-1 | P. polymyxa ATCC 842T | P. ottowii MS2379T | P. kribbensis AM49T |
|---|---|---|---|---|
| pH | 6-8 | ND | 6-10 | 5-8 |
| 温度 Temperature (℃) | 10-55 | ND | 10-45 | 10-44 |
| NaCl耐受 NaCl tolerance (%) | 5 | 2 | 3 | 4 |
| 硝酸盐还原 Nitrate reduction | + | + | + | + |
| 过氧化氢酶 Catalase | + | ND | + | - |
| 淀粉酶 Amylase | + | + | + | + |
| 吐温80 Tween 80 | + | - | ND | + |
| 牛奶胨化 Milk peptonization | + | + | + | + |
| 明胶液化 Gelatin liquefaction | + | + | + | + |
| 脲酶 Urease | - | - | - | - |
表3 菌株QH-1与其最高相似性菌株的生理生化特征比较
Table 3 Comparison of physiological and biochemical characteristics between QH-1 and its most similar strains
| 特征Characteristic | QH-1 | P. polymyxa ATCC 842T | P. ottowii MS2379T | P. kribbensis AM49T |
|---|---|---|---|---|
| pH | 6-8 | ND | 6-10 | 5-8 |
| 温度 Temperature (℃) | 10-55 | ND | 10-45 | 10-44 |
| NaCl耐受 NaCl tolerance (%) | 5 | 2 | 3 | 4 |
| 硝酸盐还原 Nitrate reduction | + | + | + | + |
| 过氧化氢酶 Catalase | + | ND | + | - |
| 淀粉酶 Amylase | + | + | + | + |
| 吐温80 Tween 80 | + | - | ND | + |
| 牛奶胨化 Milk peptonization | + | + | + | + |
| 明胶液化 Gelatin liquefaction | + | + | + | + |
| 脲酶 Urease | - | - | - | - |
| Item | QH-1-P. polymyxa ATCC 842T | QH-1-P. ottowii MS2379T | QH-1-P. kribbensis AM49T |
|---|---|---|---|
| dDDH (%) | 84.5 | 72.1 | 50.1 |
| ANI (%) | 97.98 | 91.73 | 85.26 |
表4 QH-1与其相似性菌株分子杂交和平均核苷酸鉴定分析
Table 4 dDDH and ANI of QH-1 and its similar strains
| Item | QH-1-P. polymyxa ATCC 842T | QH-1-P. ottowii MS2379T | QH-1-P. kribbensis AM49T |
|---|---|---|---|
| dDDH (%) | 84.5 | 72.1 | 50.1 |
| ANI (%) | 97.98 | 91.73 | 85.26 |
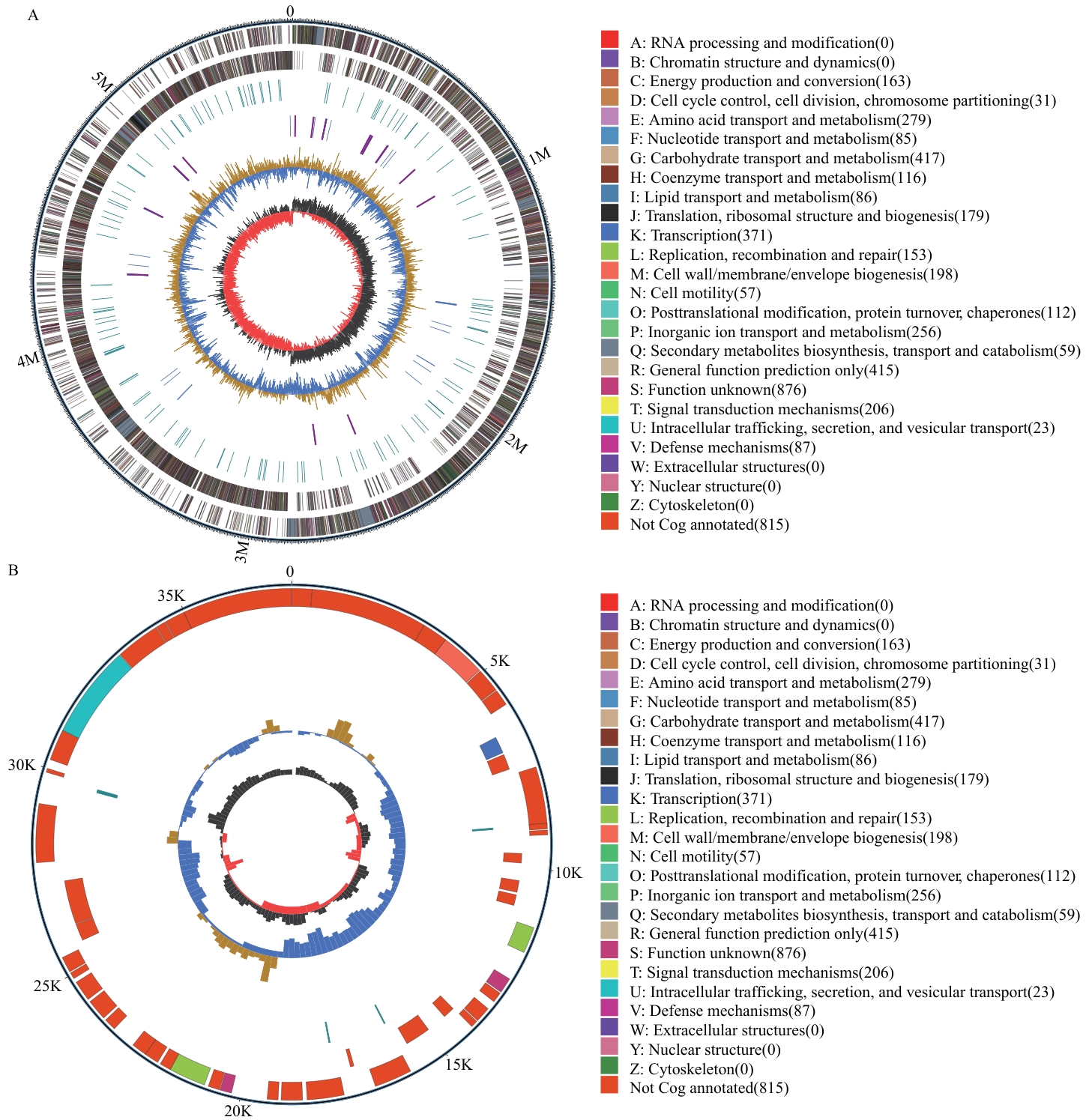
图5 QH-1的基因组圈图A:染色体圈图;B:质粒圈图。最外面一圈为基因组大小的标识;第二圈和第三圈为正链、负链上的CDS;第四圈为重复序列; 第五圈为tRNA和rRNA;第六圈为GC含量;最内一圈为GC-Skew值
Fig. 5 Genomic circle diagram of QH-1A: Chromosome circular map. B: Plasmid circular map. The outermost circle is the genome size. The second and third circles are the CDS on the positive and negative strands. The fourth circle is repeat sequences. The fifth circle is rRNA and tRNA. The sixth circle is the GC content. The innermost circle is GC-skew value
编号 Number | 基因编号 Gene number | 酶定义 Enzyme definition | 家族 Family |
|---|---|---|---|
| 1 | GE000403 | Chitinase EC 3.2.1.14 | GH18 |
| 2 | GE001510 | Chitinase EC 3.2.1.14 | GH18 |
| 3 | GE004051 | Chitinase EC 3.2.1.14 | GH18 |
| 4 | GE000622 | Cellulase EC 3.2.1.4 | GH5 |
| 5 | GE000624 | Cellulase EC 3.2.1.4 | GH5 |
| 6 | GE002468 | Endo-β-1,4-mannosidase EC 3.2.1.78 | GH5 |
| 7 | GE003415 | β-galactosidase EC 3.2.1.23 | GH5 |
| 8 | GE003585 | Endoglucanase EC 3.2.1.4 | GH5 |
| 9 | GE004895 | Cellulase EC 3.2.1.4 | GH5 |
| 10 | GE001031 | Endoglucanase EC 3.2.1.4 | GH6 |
| 11 | GE000656 | Glucosylceramidase EC 3.2.1.45 | GH30 |
| 12 | GE000930 | Glucosylceramidase EC 3.2.1.45 | GH30 |
| 13 | GE004379 | Endo-β-1,4-mannosidase EC 3.2.1.78 | GH44GH26 |
| 14 | GE002307 | Cellulase EC 3.2.1.4 | GH48 |
| 15 | GE002395 | Endo-β-(1,3)(1,4)-glucanase EC 3.2.1.6 | GH16 |
| 16 | GE004210 | Endo-β-1,3-glucanase EC 3.2.1.39 | GH81 |
表5 菌株QH-1基因组中降解细胞结构成分相关的酶
Table 5 Enzymes related to cell structural component degradation in the genome of strain QH-1
编号 Number | 基因编号 Gene number | 酶定义 Enzyme definition | 家族 Family |
|---|---|---|---|
| 1 | GE000403 | Chitinase EC 3.2.1.14 | GH18 |
| 2 | GE001510 | Chitinase EC 3.2.1.14 | GH18 |
| 3 | GE004051 | Chitinase EC 3.2.1.14 | GH18 |
| 4 | GE000622 | Cellulase EC 3.2.1.4 | GH5 |
| 5 | GE000624 | Cellulase EC 3.2.1.4 | GH5 |
| 6 | GE002468 | Endo-β-1,4-mannosidase EC 3.2.1.78 | GH5 |
| 7 | GE003415 | β-galactosidase EC 3.2.1.23 | GH5 |
| 8 | GE003585 | Endoglucanase EC 3.2.1.4 | GH5 |
| 9 | GE004895 | Cellulase EC 3.2.1.4 | GH5 |
| 10 | GE001031 | Endoglucanase EC 3.2.1.4 | GH6 |
| 11 | GE000656 | Glucosylceramidase EC 3.2.1.45 | GH30 |
| 12 | GE000930 | Glucosylceramidase EC 3.2.1.45 | GH30 |
| 13 | GE004379 | Endo-β-1,4-mannosidase EC 3.2.1.78 | GH44GH26 |
| 14 | GE002307 | Cellulase EC 3.2.1.4 | GH48 |
| 15 | GE002395 | Endo-β-(1,3)(1,4)-glucanase EC 3.2.1.6 | GH16 |
| 16 | GE004210 | Endo-β-1,3-glucanase EC 3.2.1.39 | GH81 |
基因簇编号 Cluster ID | 基因簇类型 Type | 起始位置 Start | 终止位置 End | 已知基因簇 Similar cluster | 相似度 Similarity (%) | 基因数量 Gene number |
|---|---|---|---|---|---|---|
| C1 | NRPS | 67511 | 131240 | Fusaricidin B | 100 | 34 |
| C2 | Ranthipeptide | 388821 | 410242 | - | - | 12 |
| C3 | Lanthipeptide-class-i, Cyclic-lactone-autoinducer | 1050199 | 1077551 | Paenibacillin | 100 | 22 |
| C4 | Transat-pks NRPS | 1092823 | 1170216 | - | - | 42 |
| C5 | Proteusin | 1283266 | 1303502 | - | - | 19 |
| C6 | NRPS Transat-pks | 1333876 | 1433854 | - | - | 50 |
| C7 | Lassopeptide | 1470252 | 1494319 | - | - | 23 |
| C8 | Lanthipeptide-class-i | 1777757 | 1804766 | Paenilan | 100 | 22 |
| C9 | NRPS-like, Cyclic-lactone-autoinducer | 2091488 | 2153613 | - | - | 54 |
| C10 | NRPS | 2528543 | 2622242 | Tridecaptin | 100 | 38 |
| C11 | NRPS, T1PKS | 2809874 | 2886013 | Laterocidine | 5 | 38 |
| C12 | Betalactone | 2965408 | 2996287 | Varlaxin 1046A/Varlaxin 1022A | 9 | 29 |
| C13 | Cyclic-lactone-autoinducer | 3037002 | 3057550 | - | - | 20 |
| C14 | Transat-pks NRPS, T3PKS, PKS-like | 3563899 | 3665823 | Aurantinin B/aurantinin C/aurantinin D | 35 | 55 |
| C15 | Lanthipeptide | 4368109 | 4393496 | - | - | 16 |
| C16 | NRPS | 4886738 | 4966292 | Polymyxin | 100 | 33 |
| C17 | Lanthipeptide-class-i | 5378895 | 5405329 | Paenicidin A | 100 | 19 |
表6 菌株QH-1次级代谢产物合成基因簇预测结果
Table 6 Predicted results of gene clusters for synthesis of secondary metabolites of strain QH-1
基因簇编号 Cluster ID | 基因簇类型 Type | 起始位置 Start | 终止位置 End | 已知基因簇 Similar cluster | 相似度 Similarity (%) | 基因数量 Gene number |
|---|---|---|---|---|---|---|
| C1 | NRPS | 67511 | 131240 | Fusaricidin B | 100 | 34 |
| C2 | Ranthipeptide | 388821 | 410242 | - | - | 12 |
| C3 | Lanthipeptide-class-i, Cyclic-lactone-autoinducer | 1050199 | 1077551 | Paenibacillin | 100 | 22 |
| C4 | Transat-pks NRPS | 1092823 | 1170216 | - | - | 42 |
| C5 | Proteusin | 1283266 | 1303502 | - | - | 19 |
| C6 | NRPS Transat-pks | 1333876 | 1433854 | - | - | 50 |
| C7 | Lassopeptide | 1470252 | 1494319 | - | - | 23 |
| C8 | Lanthipeptide-class-i | 1777757 | 1804766 | Paenilan | 100 | 22 |
| C9 | NRPS-like, Cyclic-lactone-autoinducer | 2091488 | 2153613 | - | - | 54 |
| C10 | NRPS | 2528543 | 2622242 | Tridecaptin | 100 | 38 |
| C11 | NRPS, T1PKS | 2809874 | 2886013 | Laterocidine | 5 | 38 |
| C12 | Betalactone | 2965408 | 2996287 | Varlaxin 1046A/Varlaxin 1022A | 9 | 29 |
| C13 | Cyclic-lactone-autoinducer | 3037002 | 3057550 | - | - | 20 |
| C14 | Transat-pks NRPS, T3PKS, PKS-like | 3563899 | 3665823 | Aurantinin B/aurantinin C/aurantinin D | 35 | 55 |
| C15 | Lanthipeptide | 4368109 | 4393496 | - | - | 16 |
| C16 | NRPS | 4886738 | 4966292 | Polymyxin | 100 | 33 |
| C17 | Lanthipeptide-class-i | 5378895 | 5405329 | Paenicidin A | 100 | 19 |
| [25] | Liu HW, Li JY, Carvalhais LC, et al. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens [J]. New Phytol, 2021, 229(5): 2873-2885. |
| [26] | Gao M, Xiong C, Gao C, et al. Disease-induced changes in plant microbiome assembly and functional adaptation [J]. Microbiome, 2021, 9(1): 187. |
| [27] | Yuan HB, Yuan MJ, Shi BK, et al. Biocontrol activity and action mechanism of Paenibacillus polymyxa strain Nl4 against pear Valsa canker caused by Valsa pyri [J]. Front Microbiol, 2022, 13: 950742. |
| [28] | Taheri E, Tarighi S, Taheri P. Characterization of root endophytic Paenibacillus polymyxa isolates with biocontrol activity against Xanthomonas translucens and Fusarium graminearum [J]. Biol Control, 2022, 174: 105031. |
| [29] | Weselowski B, Nathoo N, Eastman AW, et al. Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production [J]. BMC Microbiol, 2016, 16(1): 244. |
| [30] | Jung WJ, An KN, Jin YL, et al. Biological control of damping-off caused by Rhizoctonia solani using chitinase-producing Paenibacillus illinoisensis KJA-424 [J]. Soil Biol Biochem, 2003, 35(9): 1261-1264. |
| [31] | Zhai Y, Zhu JX, Tan TM, et al. Isolation and characterization of antagonistic Paenibacillus polymyxa HX-140 and its biocontrol potential against Fusarium wilt of cucumber seedlings [J]. BMC Microbiol, 2021, 21(1): 75. |
| [32] | Liu Y, Wang RH, Cao YH, et al. Identification and antagonistic activity of endophytic bacterial strain Paenibacillus sp. 5L8 isolated from the seeds of maize (Zea mays L., Jingke 968) [J]. Ann Microbiol, 2016, 66(2): 653-660. |
| [33] | Xu SJ, Kim BS. Evaluation of Paenibacillus polymyxa strain SC09-21 for biocontrol of Phytophthora blight and growth stimulation in pepper plants [J]. Trop Plant Pathol, 2016, 41(3): 162-168. |
| [1] | 国家药典委员会. 中华人民共和国药典-一部2015年版 [M]. 北京: 中国医药科技出版社, 2015. |
| National Pharmacopoeia Committee. People’s republic of China (PRC) pharmacopoeia-No.1 department [M]. Beijing: China Medical Science Press, 2015. | |
| [2] | Bian ZY, Zhang R, Zhang X, et al. Extraction, structure and bioactivities of polysaccharides from Rehmannia glutinosa: a review [J]. J Ethnopharmacol, 2023, 305: 116132. |
| [3] | 吴廷娟, 杜权, 王玉星, 等. 地黄根腐病病原菌的分离鉴定及其防治 [J]. 江苏农业科学, 2021, 49(22): 132-136. |
| Wu TJ, Du Q, Wang YX, et al. Isolation, identification and control of root rot pathogen of Rehmannia glutinosa [J]. Jiangsu Agric Sci, 2021, 49(22): 132-136. | |
| [4] | Lim JD, Yun SJ, Chung IM, et al. Resveratrol synthase transgene expression and accumulation of resveratrol glycoside in Rehmannia glutinosa [J]. Mol Breed, 2005, 16(3): 219-233. |
| [5] | Wu LK, Wang JY, Huang WM, et al. Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture [J]. Sci Rep, 2015, 5: 15871. |
| [6] | Cao LL, Kang QY, Tian Y. Pesticide residues: Bridging the gap between environmental exposure and chronic disease through omics [J]. Ecotoxicol Environ Saf, 2024, 287: 117335. |
| [7] | Mei LC, Chen HM, Dong AY, et al. Pesticide informatics platform (PIP): an international platform for pesticide discovery, residue, and risk evaluation [J]. J Agric Food Chem, 2022, 70(22): 6617-6623. |
| [8] | Pan XY, Yue Y, Zhao FJ, et al. Rhizosphere microbes facilitate the break of chlamydospore dormancy and root colonization of rice false smut fungi [J]. Cell Host Microbe, 2025, 33(5): 731-744.e5. |
| [9] | Wang X, Xu Q, Hu K, et al. A coculture of Enterobacter and Comamonas species reduces cadmium accumulation in rice [J]. Mol Plant Microbe Interactions® , 2023, 36(2): 95-108. |
| [10] | Wang LW, Zhang YB, Wang Y, et al. Inoculation with Penicillium citrinum aids ginseng in resisting Fusarium oxysporum by regulating the root and rhizosphere microbial communities [J]. Rhizosphere, 2022, 22: 100535. |
| [11] | Kamoun S, Furzer O, Jones JDG, et al. The Top 10 oomycete pathogens in molecular plant pathology [J]. Mol Plant Pathol, 2015, 16(4): 413-434. |
| [12] | 李文均, 刘兰, 焦建宇, 等. 细菌古菌系统分类学原理及鉴定技术 [M]. 北京: 高等教育出版社, 2024. |
| Li WJ, Liu L, Jiao JY, et al. Systematic taxonomy principle and identification technology of bacteria and archaea [M]. Beijing: Higher Education Press, 2024. | |
| [13] | He HR, Huang JR, Zhao ZZ, et al. Whole genome analysis of Streptomyces sp. RerS4, a Rehmannia glutinosa rhizosphere microbe producing a new lipopeptide [J]. Heliyon, 2023, 9(9): e19543. |
| [14] | 郭芳芳, 谢镇, 卢鹏, 等. 一株多粘类芽胞杆菌的鉴定及其生防促生效果初步测定 [J]. 中国生物防治学报, 2014, 30(4): 489-496. |
| Guo FF, Xie Z, Lu P, et al. Identification of a novel Paenibacillus polymyxa strain and its biocontrol and plant growth-promoting effects [J]. Chin J Biol Control, 2014, 30(4): 489-496. | |
| [15] | 杨剑锋, 张园园, 王娜, 等. 苜蓿根腐病病原菌的分离鉴定及苜蓿品种的抗性评价 [J]. 中国草地学报, 2020, 42(3): 52-60. |
| Yang JF, Zhang YY, Wang N, et al. Isolation and identification of pathogens causing root rot disease in alfalfa and the evaluation of resistant ability of alfalfa varieties to Fusarium equiseti and F. tricinctum [J]. Chin J Grassland, 2020, 42(3): 52-60. | |
| [16] | 陈逢玲, 孙卓, 林红梅, 等. 关防风根腐病拮抗细菌筛选与鉴定 [J]. 微生物学通报, 2022, 49(8): 3192-3204. |
| Chen FL, Sun Z, Lin HM, et al. Screening and identification of the antagonistic bacteria against root rot of Saposhnikovia divaricata [J]. Microbiol China, 2022, 49(8): 3192-3204. | |
| [17] | 黄家锐, 杜鹏强, 李文均, 等. 地黄内生放线菌的分离及地黄轮纹病拮抗菌Streptomyces folium leaf-16的新种鉴定 [J]. 微生物学报, 2022, 62(12): 4953-4963. |
| Huang JR, Du PQ, Li WJ, et al. Isolation of endophytic actinomycetes from Rehmannia glutinosa and identification of a new species Streptomyces folium leaf-16 resistant to Phoma herbarum [J]. Acta Microbiol Sin, 2022, 62(12): 4953-4963. | |
| [18] | Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees [J]. Mol Biol Evol, 1987, 4(4): 406-425. |
| [19] | Blin K, Shaw S, Kloosterman AM, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities [J]. Nucleic Acids Res, 2021, 49(W1): W29-W35. |
| [20] | Liu K, Newman M, McInroy JA, et al. Selection and assessment of plant growth-promoting rhizobacteria for biological control of multiple plant diseases [J]. Phytopathology® , 2017, 107(8): 928-936. |
| [21] | Dobrzyński J, Naziębło A. Paenibacillus as a biocontrol agent for fungal phytopathogens: is P. polymyxa the only one worth attention? [J]. Microb Ecol, 2024, 87(1): 134. |
| [22] | Luo CH, He YJ, Chen YP. Rhizosphere microbiome regulation: Unlocking the potential for plant growth [J]. Curr Res Microb Sci, 2025, 8: 100322. |
| [23] | Liu HW, Brettell LE, Qiu ZG, et al. Microbiome-mediated stress resistance in plants [J]. Trends Plant Sci, 2020, 25(8): 733-743. |
| [24] | Wen T, Zhao ML, Yuan J, et al. Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes [J]. Soil Ecol Lett, 2021, 3(1): 42-51. |
| [1] | 吕镇, 甘恬, 霍思羽, 赵晨笛, 赵梦瑶, 李亚涛, 马玉超, 耿玉清. 产Surfactin贝莱斯芽胞杆菌C5A-1的鉴定和所产Surfactin对植物的促生效果[J]. 生物技术通报, 2025, 41(9): 265-276. |
| [2] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [3] | 李亚涛, 张志鹏, 赵梦瑶, 吕镇, 甘恬, 魏浩, 吴书凤, 马玉超. 根瘤菌Bd1的全基因组分析及TetR3对细胞生长和结瘤的负调控功能[J]. 生物技术通报, 2025, 41(9): 289-301. |
| [4] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [5] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [6] | 吴泽银, 黄晨阳, 赵梦然, 张利姣, 姚方杰. 短柄白黄侧耳CCMSSC 04611基因组特异性分析[J]. 生物技术通报, 2025, 41(5): 320-332. |
| [7] | 张慧, 卢文才, 王冬, 刘倩, 马连杰. 一株高产吲哚乙酸的Bacillus cereus YT2-1C的鉴定及促生作用[J]. 生物技术通报, 2025, 41(5): 300-309. |
| [8] | 孙天国, 衣兰, 秦旭洋, 乔梦雪, 谷新颖, 韩艺, 沙伟, 张梅娟, 马天意. 大白菜DABB基因家族的全基因组鉴定及盐碱胁迫下的表达分析[J]. 生物技术通报, 2025, 41(4): 156-165. |
| [9] | 宋姝熠, 蒋开秀, 刘欢艳, 黄亚成, 刘林娅. ‘红阳’猕猴桃TCP基因家族鉴定及其在果实中的表达分析[J]. 生物技术通报, 2025, 41(3): 190-201. |
| [10] | 车建美, 郑雪芳, 王阶平, 陈燕萍, 陈冰星, 刘波. 一株产纤维素酶菌株的筛选、鉴定及全基因组分析[J]. 生物技术通报, 2025, 41(3): 294-307. |
| [11] | 宋英培, 王灿, 周会汶, 孔可可, 许孟歌, 王瑞凯. 基于全基因组关联分析和遗传多样性的大豆裂荚性状解析[J]. 生物技术通报, 2025, 41(2): 97-106. |
| [12] | 徐嘉妤, 赵阁, 唐叶, 刘雯雯, 彭清忠, 吴家和. 陆地棉WRKY基因家族全基因组鉴定及WRKY44在纤维发育中的表达分析[J]. 生物技术通报, 2025, 41(12): 124-138. |
| [13] | 苗昊翔, 张颖, 郭世鹏, 张健. 一株高产γ-氨基丁酸短乳杆菌TCCC13007全基因组测序及比较基因组分析[J]. 生物技术通报, 2025, 41(11): 166-176. |
| [14] | 李粘前, 李琛, 李淑婷, 马菊花, 景海青, 孙岩, 周雅莉, 薛金爱, 李润植. 油莎豆苹果酸酶(ME)全基因组鉴定及CeNAD-ME2功能分析[J]. 生物技术通报, 2025, 41(10): 210-221. |
| [15] | 何财林, 卢晶, 郭会会, 李小安, 吴琪. 藜麦MADS-box基因家族的全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(1): 157-172. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||