








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 95-105.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0496
胡万可1,2( ), 陈雲霞1,3, 罗帝洲4, 吴斯宇1, 李建波1, 翟少伦5, 巨向红2, 廖明6, 魏文康1(
), 陈雲霞1,3, 罗帝洲4, 吴斯宇1, 李建波1, 翟少伦5, 巨向红2, 廖明6, 魏文康1( ), 余界石1(
), 余界石1( )
)
收稿日期:2025-05-14
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
余界石,男,博士,副研究员,研究方向 :病毒学;E-mail: yujieshi@gdaas.cn作者简介:胡万可,女,研究方向 :兽医病毒学;E-mail: 2112204099@stu.gdou.edu.cn基金资助:
HU Wan-ke1,2( ), CHEN Yun-xia1,3, LUO Di-zhou4, WU Si-yu1, LI Jian-bo1, ZHAI Shao-lun5, JU Xiang-hong2, LIAO Ming6, WEI Wen-kang1(
), CHEN Yun-xia1,3, LUO Di-zhou4, WU Si-yu1, LI Jian-bo1, ZHAI Shao-lun5, JU Xiang-hong2, LIAO Ming6, WEI Wen-kang1( ), YU Jie-shi1(
), YU Jie-shi1( )
)
Received:2025-05-14
Published:2025-12-26
Online:2026-01-06
摘要:
目的 当前中国牛群中流行的丁型流感病毒多属D/Yama2019遗传进化谱系,构建此遗传进化谱系丁型流感病毒的反向遗传操作系统,为深入探究其复制与致病机制提供研究工具。 方法 以中国丁型流感病毒株D/bovine/CHN/JY3002/2022(简称D/JY3002,属D/Yama2019遗传进化谱系)为研究材料,将编码该病毒主要抗原血凝素-酯酶-融合蛋白(hemagglutinin-esterase-fusion, HEF)的基因组节段对应的DNA片段,无缝克隆至经设计改造的双向表达载体pCC1-DualPro中;其余基因组节段对应的DNA片段,则无缝克隆至常用双向表达载体pHW2000中。随后经改进优化的操作步骤,拯救出重组丁型流感病毒。 结果 拯救的丁型流感病毒株(rD/JY3002)与天然分离的中国丁型流感病毒株(D/JY3002)在复制能力上表现相当,且二者复制动力学特征相仿。rD/JY3002至少可稳定传代5代。利用已建立的反向遗传操作系统,拯救出携带绿色荧光报告基因GFP以及其他遗传进化谱系(D/OK和D/660)丁型流感病毒HEF基因的重组丁型流感病毒,分别命名为rD/JY3002-GFP、rD/JY3002-D/OK-HEF和rD/JY3002-D/660-HEF,证实了不同遗传进化谱系的丁型流感病毒间可发生基因组节段重配。 结论 建立了高效、稳定的中国丁型流感病毒株反向遗传操作系统,该系统可用于进一步开发以丁型流感病毒为载体的外源基因呈递技术。
胡万可, 陈雲霞, 罗帝洲, 吴斯宇, 李建波, 翟少伦, 巨向红, 廖明, 魏文康, 余界石. 中国丁型流感病毒株D/JY3002反向遗传操作系统的构建与功能验证[J]. 生物技术通报, 2025, 41(12): 95-105.
HU Wan-ke, CHEN Yun-xia, LUO Di-zhou, WU Si-yu, LI Jian-bo, ZHAI Shao-lun, JU Xiang-hong, LIAO Ming, WEI Wen-kang, YU Jie-shi. Establishment and Functional Validation of a Reverse Genetics System for the Chinese Influenza D Virus D/JY3002[J]. Biotechnology Bulletin, 2025, 41(12): 95-105.

图1 丁型流感病毒基因组节段5′和3′末端序列图A-G依次为不同遗传进化谱系(D/OK、D/660和D/Yama2019)丁型流感病毒PB2、 PB1、 P3、 HEF、 NP、 M和NS基因组节段5'端和3′端各100 bp序列。序列比对图中第1和2行分别是已公开的D/OK和D/660遗传进化谱系代表性毒株的基因组节段两末端序列。第3行显示的是已公开但不完整的D/JY3002基因组节段两末端序列。第4行为本研究获得的完整的D/JY3002基因组节段两末端序列。图示红框部分为丁型流感病基因组节段两末端非编码区序列
Fig. 1 5′-and 3′-end sequences of genomic segments of influenza D virusesA-G illustrate 100 bp sequences at 5'- and 3'-end of genomic segments (PB2, PB1, P3, HEF, NP, M, and NS) of influenza D viruses that belong to different genetic lineages (D/OK, D/660, and D/Yama2019). The first and second rows in each sequence alignment diagram refer to terminal sequences of genomic segments from the D/OK and D/660 strains, respectively. The third row displays the terminal sequences of the D/JY3002 genomic segment, which have been reported but are incomplete. The fourth row presents the complete terminal sequence of the D/JY3002 genomic segment obtained in this study. The red box in the illustration highlights the sequences of the non-coding regions at both ends of genomic segments of influenza D viruses
| Number | Primer name | Primer sequence(5'-3') |
|---|---|---|
| 1 | D/JY3002-PB2-F | AGCATAAGCAGAGGATGTCACTACTATTAACGC |
| 2 | D/JY3002-PB2-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCAATGTG |
| 3 | D/JY3002-PB1-F | GGAGCATAAGCAGAGGATTTTATAACAATGGA |
| 4 | D/JY3002-PB1-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTC |
| 5 | D/JY3002-P3-F | AGCATAAGCAGGAGATTTAGAAATGTCTAGTAT |
| 6 | D/JY3002-P3-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTAA |
| 7 | D/JY3002-HEF-F | AGCATAAGCAGGAGATTTTCAAAGATGTTTTTG |
| 8 | D/JY3002-HEF-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTCTAAGAT |
| 9 | D/JY3002-NP-F | AGCATAAGCAGGAGATTATTAAGCAATATGGAC |
| 10 | D/JY3002-NP-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTGTTAAAT |
| 11 | D/JY3002-M-F | GGAGCATAAGCAGAGGATATTTTTGACGCAATG |
| 12 | D/JY3002-M-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCGCGA |
| 13 | D/JY3002-NS-F | GGAGCATAAGCAGGGGTGTACAATTTCAATATG |
| 14 | D/JY3002-NS-R | CCGCCGGGTTATTAGCAGTAGCAAGGGGTTTTTTCATACT |
| 15 | pHW2000-F | TACTGCTAATAACCCGGCGGCCCAAAATGCCG |
| 16 | pHW2000-PB2-R | TGACATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 17 | pHW2000-PB1-R | AAATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 18 | pHW2000-P3-R | CTAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 19 | pHW2000-HEF-R | GAAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 20 | pHW2000-NP-R | AATAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 21 | pHW2000-M-R | ATATCCTCTGCTTATGCTCCCCCCCAAACTTCGGAGGTCGA |
| 22 | pHW2000-NS-R | TACACCCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
表1 目的片段及载体线性化扩增引物
Table 1 Primers for amplifying target fragments and linearization of vectors
| Number | Primer name | Primer sequence(5'-3') |
|---|---|---|
| 1 | D/JY3002-PB2-F | AGCATAAGCAGAGGATGTCACTACTATTAACGC |
| 2 | D/JY3002-PB2-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCAATGTG |
| 3 | D/JY3002-PB1-F | GGAGCATAAGCAGAGGATTTTATAACAATGGA |
| 4 | D/JY3002-PB1-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTC |
| 5 | D/JY3002-P3-F | AGCATAAGCAGGAGATTTAGAAATGTCTAGTAT |
| 6 | D/JY3002-P3-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTAA |
| 7 | D/JY3002-HEF-F | AGCATAAGCAGGAGATTTTCAAAGATGTTTTTG |
| 8 | D/JY3002-HEF-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTCTAAGAT |
| 9 | D/JY3002-NP-F | AGCATAAGCAGGAGATTATTAAGCAATATGGAC |
| 10 | D/JY3002-NP-R | CCGCCGGGTTATTAGCAGTAGCAAGGAGATTTTTTGTTAAAT |
| 11 | D/JY3002-M-F | GGAGCATAAGCAGAGGATATTTTTGACGCAATG |
| 12 | D/JY3002-M-R | CCGCCGGGTTATTAGCAGTAGCAAGAGGATTTTTTCGCGA |
| 13 | D/JY3002-NS-F | GGAGCATAAGCAGGGGTGTACAATTTCAATATG |
| 14 | D/JY3002-NS-R | CCGCCGGGTTATTAGCAGTAGCAAGGGGTTTTTTCATACT |
| 15 | pHW2000-F | TACTGCTAATAACCCGGCGGCCCAAAATGCCG |
| 16 | pHW2000-PB2-R | TGACATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 17 | pHW2000-PB1-R | AAATCCTCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 18 | pHW2000-P3-R | CTAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 19 | pHW2000-HEF-R | GAAAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 20 | pHW2000-NP-R | AATAATCTCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
| 21 | pHW2000-M-R | ATATCCTCTGCTTATGCTCCCCCCCAAACTTCGGAGGTCGA |
| 22 | pHW2000-NS-R | TACACCCCTGCTTATGCTCCCCCCCAACTTCGGAGGTCGA |
图2 构建含有D/JY3002基因组节段对应DNA片段的双向表达质粒A:RT-PCR扩增获得D/JY3002全长基因组节段PB2(2 364 bp)、PB1(2 330 bp)、P3(2 195 bp)、HEF(2 049 bp)、NP(1 775 bp)、M(1 219 bp) 和 NS(868 bp)对应的DNA片段的凝胶电泳图;B:双向表达质粒pHW2000连接了D/JY3002基因组节段PB2、PB1、P3、NP、M或NS对应DNA片段的图谱示意图;C:将D/JY3002 HEF基因组节段对应的DNA片段无缝至pHW2000载体。对转化后获得的菌落进行PCR鉴定和凝胶电泳分析,其中泳道1-7分别代表7个不同的菌落样品;D:设计改造的双向表达质粒pCC1-DualPro连接了D/JY3002基因组节段HEF对应DNA片段的图谱示意图;E:将D/JY3002 HEF基因组节段对应的DNA片段,通过无缝克隆技术连接至pCC1-DualPro载体。对转化后获得的菌落进行PCR鉴定和凝胶电泳分析,其中泳道1-2分别代表2个不同的菌落样品;F:对提取的含有D/JY3002基因组节段对应DNA片段的所有双向表达质粒进行凝胶电泳分析,泳道1-7分别为pHW2000-D/JY3002-PB2 (5 377 bp)、-PB1 (5 343 bp)、-P3 (5 208 bp)、-NP (4 788 bp)、-M (4 232 bp)、-NS (3 881 bp),和pCC1-DualPro-D/JY3002-HEF (11 105 bp)。Marker,用于指示DNA片段的大小
Fig. 2 Construction of bidirectional expression plasmids containing DNA fragments corresponding to the D/JY3002 genomic segmentsA: Gel electrophoresis plots of amplified DNA fragments corresponding to the full-length genomic segments PB2 (2 364 bp), PB1 (2 330 bp), P3 (2 195 bp), HEF (2 049 bp), NP (1 775 bp), M (1 219 bp) and NS (868 bp) of the D/JY3002. B: The map of bidirectional plasmid pHW2000 containing the DNA fragment corresponding to the full-length genomic segment PB2, PB1, P3, NP, M or NS of the D/JY3002.C: The DNA fragment corresponding to the D/JY3002 HEF genomic segment was ligated to the pHW2000 vector through seamless cloning technology. The colonies obtained after transformation were identified by PCR and analyzed by gel electrophoresis. Among them, lane 1 to 7 indicate 7 different colony samples respectively. D: The map of designed bidirectional plasmid pCC1-DualPro containing the DNA fragment corresponding to the full-length genomic segment HEF of the D/JY3002. E: The DNA fragment corresponding to the D/JY3002 HEF genomic segment was ligated to the pCC1-DualPro vector through seamless cloning technology. The colonies obtained after transformation were identified by PCR and analyzed by gel electrophoresis. Among them, lane 1 and 2 indicate 2 different colony samples. F: Gel electrophoresis analysis was performed on all the extracted bidirectional expression plasmids. Lane 1 to 7 are pHW2000-D/JY3002-PB2 (5 377 bp), -PB1 (5 343 bp), -P3 (5 208 bp), -NP (4 788 bp), -M (4 232 bp), -NS (3 881 bp), and pCC1-DualPro-D/JY3002-HEF (11 105 bp), respectively. Marker, used to indicate the size of the DNA fragment
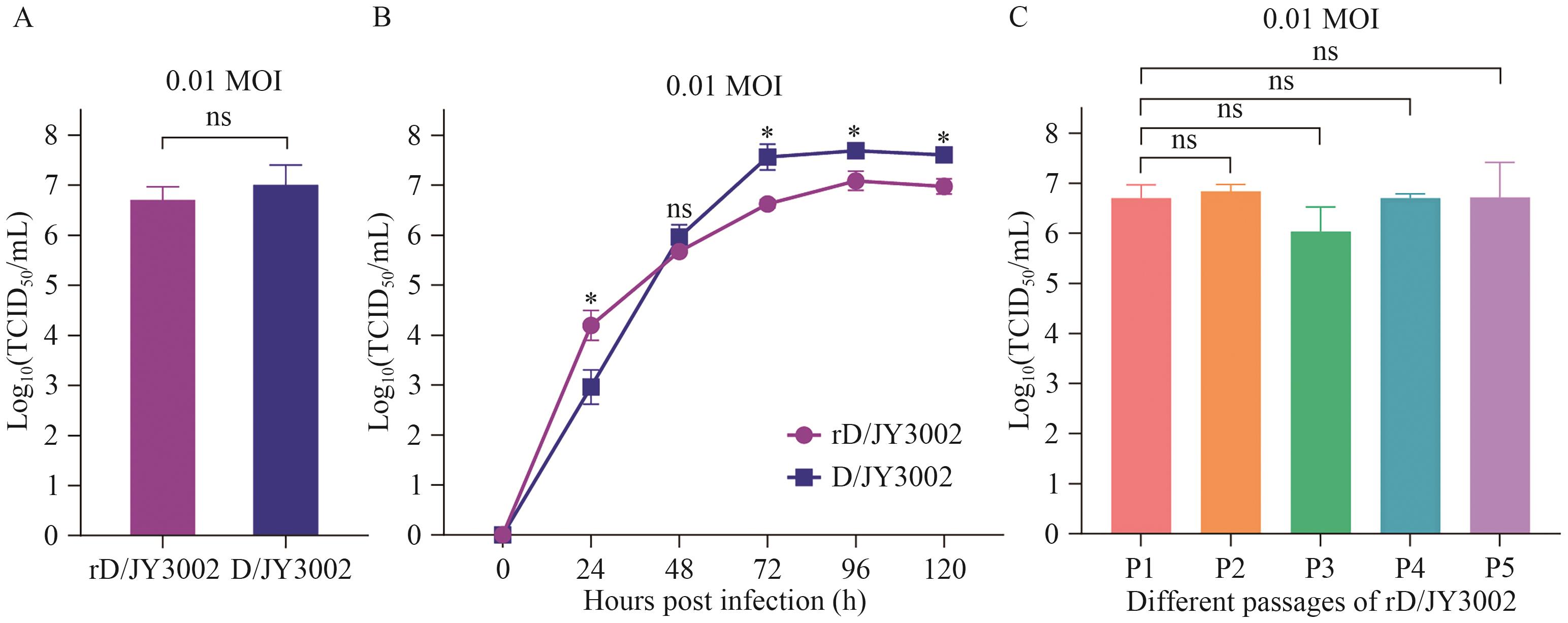
图3 人工拯救rD/JY3002与野生D/JY3002丁型流感病毒复制能力和生长动力学对比分析A:人工拯救病毒rD/JY3002与自然分离病毒D/JY3002的复制滴度(TCID50/mL);B:rD/JY3002与D/JY3002的生长曲线;C:不同代次rD/JY3002(P1、P2、P3、P4和P5)的复制滴度。感染剂量为0.01 MOI, “ns”表示无显著差异, *:P<0.05;**:P<0.01;***:P<0.001
Fig. 3 Comparative analysis of the replication capacity and growth kinetics of artificially rescued rD/JY3002 and wild-type D/JY3002 influenza D virusesA: Replication titers (TCID50/mL) of the rescued rD/JY3002 and naturally isolated D/JY3002. B: Growth curves of rD/JY3002 and D/JY3002. C: Replication titers of different passages of rD/JY3002 (P1, P2, P3, P4 and P5). The infection dose was 0.01 MOI, and "ns" indicates no significant difference. * : P<0.05; ** : P<0.01 *** : P<0.001
图4 rD/JY3002-GFP的人工拯救与GFP表达分析A:设计构建单向表达质粒pPolI-D/JY3002-PB1-240-GFP-240(图谱示意图);B:不同代次人工拯救病毒rD/JY3002-GFP的复制滴度(TCID50/mL);C:不同代次人工拯救病毒rD/JY3002-GFP感染MDCK细胞后的荧光图。Mock/Infected: 未感染/感染细胞。感染剂量为0.01 MOI
Fig. 4 Artificial rescue of the rD/JY3002-GFP and expression analysis of the GFPA: Design and construction of the unidirectional expression plasmid pPolI-D/JY3002-PB1-240-GFP-240 (schematic diagram). B: Replication titers (TCID50/mL) of different passages of rD/JY3002-GFP viruses. C: Fluorescence images of MDCK cells infected with different passages of rD/JY3002-GFP viruses. Mock/Infected: Uninfected/Infected cells, and the infection dose is 0.01 MOI
图5 携带异源HEF基因的重组丁型流感病毒的构建及其抗原交叉性分析A:重组丁型流感病毒rD/JY3002、rD/JY3002-D/OK-HEF和rD/JY3002-D/660-HEF示意图;B:rD/JY3002、rD/JY3002-D/OK-HEF和rD/JY3002-D/660-HEF在MDCK细胞上的生长曲线;C:兔抗D/JY3002标准阳性血清针对重组丁型流感病毒rD/JY3002、rD/JY3002-D/OK-HEF和rD/JY3002-D/660-HEF的HI效价
Fig. 5 Construction of recombinant influenza D viruses containing the heterologous HEF gene and analysis of their antigenic cross-reactivityA: Schematic diagrams of recombinant influenza D viruses rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF. B: Growth kinetics of rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF on MDCK cells. C: HI titers of rabbit anti-D/JY3002 sera against recombinant influenza D viruses rD/JY3002, rD/JY3002-D/OK-HEF and rD/JY3002-D/660-HEF
| [1] | Hause BM, Ducatez M, Collin EA, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses [J]. PLoS Pathog, 2013, 9(2): e1003176. |
| [2] | Hause BM, Collin EA, Liu RX, et al. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae Family [J]. mBio, 2014, 5(2): e00031-14. |
| [3] | Collin EA, Sheng ZZ, Lang YK, et al. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle [J]. J Virol, 2015, 89(2): 1036-1042. |
| [4] | Ferguson L, Olivier AK, Genova S, et al. Pathogenesis of influenza D virus in cattle [J]. J Virol, 2016, 90(12): 5636-5642. |
| [5] | Yu JS, Li F, Wang D. The first decade of research advances in influenza D virus [J]. J Gen Virol, 2021, 102(1). |
| [6] | Trombetta CM, Marchi S, Marotta MG, et al. Detection of influenza D antibodies in dogs, Apulia Region, Italy, 2016 and 2023 [J]. Emerg Infect Dis, 2024, 30(5): 1045-1047. |
| [7] | Guan MH, Jacobson O, Sarafianos G, et al. Exposure of white-tailed deer in North America to influenza D virus [J]. Virology, 2022, 573: 111-117. |
| [8] | Yu JS, Hika B, Liu RX, et al. The hemagglutinin-esterase fusion glycoprotein is a primary determinant of the exceptional thermal and acid stability of influenza D virus [J]. mSphere, 2017, 2(4) |
| [9] | Huang C, Yu JS, Hause BM, et al. Emergence of new phylogenetic lineage of Influenza D virus with broad antigenicity in California, United States [J]. Emerg Microbes Infect, 2021, 10(1): 739-742. |
| [10] | Yu JS, Li TY, Wen ZY, et al. Identification of D/Yama2019 lineage-like influenza D virus in Chinese cattle [J]. Front Vet Sci, 2022, 9: 939456. |
| [11] | Yu JS, Wen ZY, Hu WK, et al. Influenza D virus infection in China, 2022-2023 [J]. Emerg Microbes Infect, 2024, 13(1): 2343907. |
| [12] | Yu JS, Liu RX, Zhou B, et al. Development and characterization of a reverse-genetics system for influenza D virus [J]. J Virol, 2019, 93(21): e01186-19. |
| [13] | Ishida H, Murakami S, Kamiki H, et al. Establishment of a reverse genetics system for influenza D virus [J]. J Virol, 2020, 94(10): e01767-19. |
| [14] | Holwerda M, Laloli L, Wider M, et al. Establishment of a reverse genetic system from a bovine derived influenza D virus isolate [J]. Viruses, 2021, 13(3): 502. |
| [15] | Murakami S, Sato R, Ishida H, et al. Influenza D virus of new phylogenetic lineage, Japan [J]. Emerg Infect Dis, 2020, 26(1): 168-171. |
| [16] | Hoffmann E, Neumann G, Kawaoka Y, et al. A DNA transfection system for generation of influenza A virus from eight plasmids [J]. Proc Natl Acad Sci U S A, 2000, 97(11): 6108-6113. |
| [17] | Hoffmann E, Mahmood K, Yang CF, et al. Rescue of influenza B virus from eight plasmids [J]. Proc Natl Acad Sci U S A, 2002, 99(17): 11411-11416. |
| [18] | Crescenzo-Chaigne B, van der Werf S. Rescue of influenza C virus from recombinant DNA [J]. J Virol, 2007, 81(20): 11282-11289. |
| [19] | Pleschka S, Jaskunas R, Engelhardt OG, et al. A plasmid-based reverse genetics system for influenza A virus [J]. J Virol, 1996, 70(6): 4188-4192. |
| [20] | Fodor E, Devenish L, Engelhardt OG, et al. Rescue of influenza a virus from recombinant DNA [J]. J Virol, 1999, 73(11): 9679-9682. |
| [21] | Noda T. Selective genome packaging mechanisms of influenza a viruses [J]. Cold Spring Harb Perspect Med, 2020: a038497. |
| [22] | Jiang WM, Wang SC, Peng C, et al. Identification of a potential novel type of influenza virus in Bovine in China [J]. Virus Genes, 2014, 49(3): 493-496. |
| [23] | Zhai SL, Zhang H, Chen SN, et al. Influenza D virus in animal species in Guangdong Province, Southern China [J]. Emerg Infect Dis, 2017, 23(8): 1392-1396. |
| [24] | He WT, Lu M, Xing G, et al. Emergence and adaptive evolution of influenza D virus [J]. Microb Pathog, 2021, 160: 105193. |
| [25] | 余界石, 魏文康. 丁型流感病毒在中国的流行状况及研究进展 [J]. 广东农业科学, 2022, 49(11): 1-8. |
| Yu JS, Wei WK. Epidemiology and research advances of influenza D virus in China [J]. Guangdong Agric Sci, 2022, 49(11): 1-8. | |
| [26] | Gao HB, Sun WY, Lu PY, et al. First isolation of influenza D virus from cattle in Northeast China [J]. Microbiol Spectr, 2024, 12(9): e00374-24. |
| [27] | Lim EH, Lim SI, Kim MJ, et al. First detection of influenza D virus infection in cattle and pigs in the republic of Korea [J]. Microorganisms, 2023, 11(7): 1751. |
| [28] | Nakatsu S, Murakami S, Shindo K, et al. Influenza C and D viruses package eight organized ribonucleoprotein complexes [J]. J Virol, 2018, 92(6): e02084-17. |
| [29] | Nakatsu S, Sagara H, Sakai-Tagawa Y, et al. Complete and incomplete genome packaging of influenza a and B viruses [J]. mBio, 2016, 7(5): e01248-16. |
| [1] | 豆硕, 丁若羲, 孙星, 郭文静, 孔文慧, 袁静贤, 张冬梅, 王省芬, 马峙英, 吴金华, 吴立柱. 一种可视化筛选转基因通用载体pCamRUBY的构建和应用研究[J]. 生物技术通报, 2025, 41(12): 66-73. |
| [2] | 刘言, 朱龙佼, 张文强, 许文涛. 生物基纳米食品工程研究进展、挑战与前景[J]. 生物技术通报, 2025, 41(11): 100-109. |
| [3] | 王静, 常雪瑞, 贾旭, 黄嘉欣, 王田田, 梁燕平. 辣椒CaUBC38基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(10): 242-252. |
| [4] | 张静安, 胡孝龙, 曹蓓蓓, 廖敏, 束长龙, 张杰, 王奎, 操海群. 苏云金芽胞杆菌可视化快速表达载体的构建与特性分析[J]. 生物技术通报, 2025, 41(1): 95-102. |
| [5] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [6] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [7] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [8] | 梅显军, 宋慧洋, 李京昊, 梅超, 宋倩娜, 冯瑞云, 陈喜明. 马铃薯StDof5的克隆及表达分析[J]. 生物技术通报, 2024, 40(3): 181-192. |
| [9] | 周家伟, 武志强. mitoTALENs植物线粒体基因编辑载体的构建方法[J]. 生物技术通报, 2024, 40(10): 172-180. |
| [10] | 李焕敏, 高峰涛, 李伟忠, 王金庆, 封佳丽. 天然生物质材料作为固定化载体的研究应用进展[J]. 生物技术通报, 2023, 39(7): 105-112. |
| [11] | 陆新华, 孙德权, 张秀梅. 介孔硅纳米粒作为植物细胞转基因载体的研究[J]. 生物技术通报, 2022, 38(7): 194-204. |
| [12] | 黎智康, 刘晨雪璇, 谭楚敏, 熊盛, 谢秋玲. MFG-E8作为外泌体载体蛋白的作用与功能[J]. 生物技术通报, 2022, 38(4): 288-294. |
| [13] | 邹雪峰, 李铭刚, 包玲风, 陈齐斌, 赵江源, 汪林, 濮永瑜, 郝大程, 张庆, 杨佩文. 一株分泌型铁载体真菌分离鉴定及生物活性研究[J]. 生物技术通报, 2022, 38(3): 130-138. |
| [14] | 刘萌萌, 韩立军, 刘宝玲, 薛金爱, 李润植. 陆地棉GhSDP1及其启动子的克隆与表达分析[J]. 生物技术通报, 2022, 38(2): 34-43. |
| [15] | 吴坤坤, 徐行, 季策, 任建峰, 李伟明, 张庆华. 斑马鱼notch3基因真核表达载体的构建及其表达分析[J]. 生物技术通报, 2022, 38(1): 179-186. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||