








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 65-71.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1068
• 食用菌生物技术专题(专题主编: 黄晨阳) • 上一篇 下一篇
收稿日期:2021-08-20
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:齐雨慧,女,硕士研究生,研究方向:食用菌遗传育种与栽培;E-mail: 基金资助:
QI Yu-hui( ), HUANG Chen-yang, ZHANG Li-jiao(
), HUANG Chen-yang, ZHANG Li-jiao( )
)
Received:2021-08-20
Published:2021-11-26
Online:2021-12-03
摘要:
转录因子XBP1是真菌发育和其他生物过程的关键调节因子。利用RNAi法探究其在糙皮侧耳生长发育过程中的功能。以糙皮侧耳CCMSSC00389为材料,克隆获得PoXBP1基因,全长1 326 bp,编码441个氨基酸,生物信息学分析结果表明XBP1蛋白含有HTH APSES结构域,无信号肽,是非跨膜结构蛋白。在野生型菌株的不同发育阶段的表达显示,PoXBP1基因在菌丝、原基、孢子、菌盖、菌柄中的表达依次降低。通过农杆菌介导的遗传转化法筛选获得PoXBP1干扰菌株,其中,干扰菌株RNAiPoXBP1-1和RNAiPoXBP1-4的生长速度慢于野生型菌株,表明了PoXBP1表达量降低,延缓了糙皮侧耳菌丝和子实体的生长速度。
齐雨慧, 黄晨阳, 张利姣. RNAi法分析糙皮侧耳XBP1转录因子[J]. 生物技术通报, 2021, 37(11): 65-71.
QI Yu-hui, HUANG Chen-yang, ZHANG Li-jiao. Analysis of XBP1 Transcription Factor in Pleurotus ostreatus by RNAi[J]. Biotechnology Bulletin, 2021, 37(11): 65-71.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose |
|---|---|---|
| PoXBP1-Spel-F1 | ATGATTGACTCACAGCCGGACT | 克隆 |
| PoXBP1-Apal-R1 | TCAGTTGAGGATATTTGACCAA | |
| PoXBP1-Spe1-R | CGACTAGTCCGTATGGAGAAT- ATCGGCTAT | |
| PoXBP1-Apa1-F | CCGGGCCCATGATTGACTCA- CAGCCGGA | |
| PoXBP1-intro-Spe1-R | GGACTAGTGGGAATATGAT- GCGCCGCTGGC | |
| PoXBP1-Bg111-F | GAAGATCTTCATGATTGAC- TCACAGCCGGA | |
| PoXBP1-qrt-SF1 | ATACAGGGAACATGGATGCC | qPCR引物 |
| PoXBP1-qrt-SR1 | ACTTGACCATAACCAGGCTG | |
| β-actin-F | AGTCGGTGCCTTGGTTAT | 内参基因 |
| β-actin-R | ATACCGACCATCACACCT | |
| hyg-F | AGAAGATGTTGGCGACCTC | 检测转化子 |
| hyg-R | AGCGAGAGCCTGACCTATTG |
表1 引物序列
Table 1 Primer sequence
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose |
|---|---|---|
| PoXBP1-Spel-F1 | ATGATTGACTCACAGCCGGACT | 克隆 |
| PoXBP1-Apal-R1 | TCAGTTGAGGATATTTGACCAA | |
| PoXBP1-Spe1-R | CGACTAGTCCGTATGGAGAAT- ATCGGCTAT | |
| PoXBP1-Apa1-F | CCGGGCCCATGATTGACTCA- CAGCCGGA | |
| PoXBP1-intro-Spe1-R | GGACTAGTGGGAATATGAT- GCGCCGCTGGC | |
| PoXBP1-Bg111-F | GAAGATCTTCATGATTGAC- TCACAGCCGGA | |
| PoXBP1-qrt-SF1 | ATACAGGGAACATGGATGCC | qPCR引物 |
| PoXBP1-qrt-SR1 | ACTTGACCATAACCAGGCTG | |
| β-actin-F | AGTCGGTGCCTTGGTTAT | 内参基因 |
| β-actin-R | ATACCGACCATCACACCT | |
| hyg-F | AGAAGATGTTGGCGACCTC | 检测转化子 |
| hyg-R | AGCGAGAGCCTGACCTATTG |
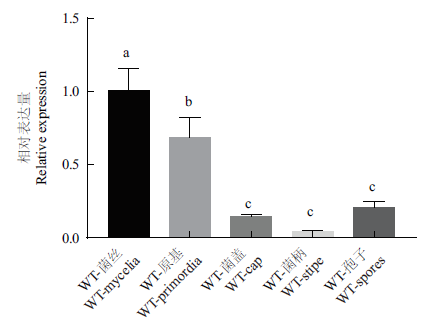
图4 野生型菌株不同组织中PoXBP1基因相对表达量比较 不同小写字母代表差异显著(P<0.05),下同
Fig.4 Comparison of the relative expression of PoXBP1 gene in different tissues of wild-type strains Different lowercase letters represent significant differences(P<0.05). The same below

图5 RNAi-PoXBP1转化子筛选 M:DL2000 marker;1-18:抗性菌块DNA;19:阳性对照:质粒RNAi-PoXBP1;20:阴性对照:389 DNA
Fig.5 Screening of RNAi-PoXBP1 transformants M:DL2000 marker;1-18:fast DNA of resistant bacteria;19:plasmid RNAi-PoXBP1;20:389 DNA
| 菌株 Strain | 菌丝顶端细胞长度 Length of mycelial apical cell/μm | 菌丝顶端细胞直径 Diameter of mycelial apical cell/μm | 菌丝顶端细胞表面积 Surface area of mycelial apical cells/μm2 |
|---|---|---|---|
| 389 | 105 ± 17a | 4.90 ± 0.95 | 500 ± 187a |
| RNAipoXBP1-4 | 72 ± 16b | 4.91 ± 1.09 | 348 ± 164b |
| RNAipoXBP1-1 | 69 ± 13b | 4.97 ± 0.88 | 321 ± 117b |
表2 菌丝显微形态对比
Table 2 Comparison of mycelial morphology
| 菌株 Strain | 菌丝顶端细胞长度 Length of mycelial apical cell/μm | 菌丝顶端细胞直径 Diameter of mycelial apical cell/μm | 菌丝顶端细胞表面积 Surface area of mycelial apical cells/μm2 |
|---|---|---|---|
| 389 | 105 ± 17a | 4.90 ± 0.95 | 500 ± 187a |
| RNAipoXBP1-4 | 72 ± 16b | 4.91 ± 1.09 | 348 ± 164b |
| RNAipoXBP1-1 | 69 ± 13b | 4.97 ± 0.88 | 321 ± 117b |
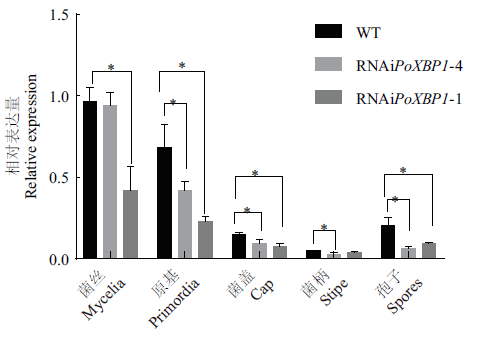
图9 干扰子与野生型不同时期、部位PoXBP1基因相对表达量比较 *代表差异显著(P<0.05)
Fig.9 Comparison of relative expressions of PoXBP1 gene in different periods and tissues between RNAi strains and wild type * represents significant difference(P<0.05)
| [1] |
Han JW, Kim DY, Lee YJ, et al. Transcription factor PdeR is involved in fungal development, metabolic change, and pathogenesis of gray mold Botrytis cinerea[J]. J Agric Food Chem, 2020, 68(34): 9171-9179.
doi: 10.1021/acs.jafc.0c02420 URL |
| [2] |
Fu H, Chung KR, Gai Y, et al. The basal transcription factor II H subunit Tfb5 is required for stress response and pathogenicity in the tangerine pathotype of Alternaria alternata[J]. Mol Plant Pathol, 2020, 21(10): 1337-1352.
doi: 10.1111/mpp.12982 URL |
| [3] |
Wang JJ, Qiu L, Cai Q, et al. Transcriptional control of fungal cell cycle and cellular events by Fkh2, a forkhead transcription factor in an insect pathogen[J]. Sci Rep, 2015, 5: 10108.
doi: 10.1038/srep10108 URL |
| [4] |
Meng L, Lyu X, Shi L, et al. The transcription factor FvHmg1 negatively regulates fruiting body development in Winter Mushroom Flammulina velutipes[J]. Gene, 2021, 785: 145618.
doi: 10.1016/j.gene.2021.145618 URL |
| [5] |
Wang W, Wang L, Chen B, et al. Characterization and expression pattern of homeobox transcription factors in fruiting body development of straw mushroom Volvariella volvacea[J]. Fungal Biol, 2019, 123(2): 95-102.
doi: 10.1016/j.funbio.2018.10.008 URL |
| [6] | 周烁红, 沈颖越, 蔡为明, 等. 肺形侧耳变温结实相关基因ppcsl-1的克隆及功能预测[J]. 菌物学报, 2016, 35(8): 946-955. |
| Zhou SH, Shen YY, Cai WM, et al. Cloning and functional prediction of the Ppcsl 1 related to change-temperature fruiting of Pleurotus pulmonarius[J]. Mycosystema, 2016, 35(8): 946-955. | |
| [7] |
Qi YC, Chen HJ, Zhang MK, et al. Identification and expression analysis of Pofst3 suggests a role during Pleurotus ostreatus primordia formation[J]. Fungal Biol, 2019, 123(3): 200-208.
doi: 10.1016/j.funbio.2018.12.008 URL |
| [8] |
Chen SY, Yang WT, Jia QM, et al. Pleurotus ostreatus bHLH transcription factors regulate plant growth and development when expressed in Arabidopsis[J]. J Plant Interact, 2017, 12(1): 542-549.
doi: 10.1080/17429145.2017.1400124 URL |
| [9] |
Zhao Y, Su H, Zhou J, et al. The APSES family proteins in fungi:Characterizations, evolution and functions[J]. Fungal Genet Biol, 2015, 81: 271-280.
doi: 10.1016/j.fgb.2014.12.003 URL |
| [10] |
Sánchez-Gaya V, Casaní-Galdón S, Ugidos M, et al. Elucidating the role of chromatin state and transcription factors on the regulation of the yeast metabolic cycle:a multi-omic integrative approach[J]. Front Genet, 2018, 9: 578.
doi: 10.3389/fgene.2018.00578 pmid: 30555512 |
| [11] |
Mai B, Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family[J]. Mol Cell Biol, 1997, 17(11): 6491-6501.
doi: 10.1128/MCB.17.11.6491 pmid: 9343412 |
| [12] | Chakraborty R, Baek JH, Bae EY, et al. Comparison and contrast of plant, yeast, and mammalian ER stress and UPR[J]. Appl Biol Chem, 2016, 59(3): 337-347. |
| [13] |
Tao R, Chen H, Gao C, et al. Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast[J]. Cell Res, 2011, 21(11): 1619-1633.
doi: 10.1038/cr.2011.58 URL |
| [14] |
Miled C, Mann C, Faye G. Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth[J]. Mol Cell Biol, 2001, 21(11): 3714-3724.
pmid: 11340165 |
| [15] |
Miles S, Li L, Davison J, et al. Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state[J]. PLoS Genet, 2013, 9(10): e1003854.
doi: 10.1371/journal.pgen.1003854 URL |
| [16] | 王丽宁. 糙皮侧耳过氧化氢酶基因特征分析和功能研究[D]. 北京:中国农业科学院, 2019. |
| Wang LN. Characterization and function analysis of catalase genes in Pleurotus ostreatus[D]. Beijing:Chinese Academy of Agricultural Sciences, 2019. | |
| [17] | 刘秀明, 邬向丽, 陈强, 等. 高温胁迫对刺芹侧耳菌丝生长及其抗棘孢木霉能力的影响[J]. 菌物学报, 2017, 36(11): 1566-1574. |
| Liu XM, Wu XL, Chen Q, et al. Effects of heat stress on Pleurotus eryngii mycelial growth and its resistance to Trichoderma asperellum[J]. Mycosystema, 2017, 36(11): 1566-1574. | |
| [18] | 侯志浩, 赵梦然, 陈强, 等. 热胁迫下糙皮侧耳实时荧光定量PCR内参基因的选择[J]. 食用菌学报, 2019, 26(3): 11-18, 157. |
| Hou ZH, Zhao MR, Chen Q, et al. Selection of reference genes for real-time quantitative PCR of Pleurotus ostreatus under heat stress[J]. Acta Edulis Fungi, 2019, 26(3): 11-18, 157. | |
| [19] | 周烁红. 秀珍菇变温结实相关ppcsl-1基因的克隆及表达分析[D]. 金华:浙江师范大学, 2016. |
| Zhou SH. Cloning and expression analysis of the ppcsl-1 gene related to change-temperature fruiting from Pleurotus pulmona-rius[D]. Jinhua:Zhejiang Normal University, 2016. | |
| [20] |
Hou L, Wang L, Wu X, et al. Expression patterns of two pal genes of Pleurotus ostreatus across developmental stages and under heat stress[J]. BMC Microbiol, 2019, 19(1): 231.
doi: 10.1186/s12866-019-1594-4 URL |
| [21] | 左勇涛. 平菇NADPH氢化酶基因NOXA的特性及功能研究[D]. 郑州:河南农业大学, 2013. |
| Zuo YT. Studies on the characteristics and function of NADPH oxidase gene NOXA in Pleurotus ostreatus[D]. Zhengzhou:Henan Agricultural University, 2013. | |
| [22] |
Borna H, Imani S, Iman M, et al. Therapeutic face of RNAi:in vivo challenges[J]. Expert Opin Biol Ther, 2015, 15(2): 269-285.
doi: 10.1517/14712598.2015.983070 URL |
| [23] |
Nakayashiki H, Hanada S, Nguyen BQ, et al. RNA silencing as a tool for exploring gene function in ascomycete fungi[J]. Fungal Genet Biol, 2005, 42(4): 275-283.
pmid: 15749047 |
| [24] |
Nguyen QB, Kadotani N, Kasahara S, et al. Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system[J]. Mol Microbiol, 2008, 68(6): 1348-1365.
doi: 10.1111/j.1365-2958.2008.06242.x pmid: 18433453 |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [7] | 胡海琳, 徐黎, 李晓旭, 王晨璨, 梅曼, 丁文静, 赵媛媛. 小肽激素调控植物生长发育及逆境生理研究进展[J]. 生物技术通报, 2023, 39(7): 13-25. |
| [8] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [9] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [10] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [11] | 薛皦, 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋. 植物基因上游开放阅读框的研究进展[J]. 生物技术通报, 2023, 39(4): 157-165. |
| [12] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [13] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [14] | 桑田, 王鹏程. 植物SUMO化修饰研究进展[J]. 生物技术通报, 2023, 39(3): 1-12. |
| [15] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||