








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 72-80.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0444
• 食用菌生物技术专题(专题主编: 黄晨阳) • 上一篇 下一篇
黄海辰1( ), 吴文雅1,2, 戚梦1,2, 薛帆正1, 吴小平1,2, 张君丽3, 傅俊生1,2(
), 吴文雅1,2, 戚梦1,2, 薛帆正1, 吴小平1,2, 张君丽3, 傅俊生1,2( )
)
收稿日期:2021-04-07
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:黄海辰,男,研究方向:食用菌活性物质研究;E-mail: 基金资助:
HUANG Hai-chen1( ), WU Wen-ya1,2, QI Meng1,2, XUE Fan-zheng1, WU Xiao-ping1,2, ZHANG Jun-li3, FU Jun-sheng1,2(
), WU Wen-ya1,2, QI Meng1,2, XUE Fan-zheng1, WU Xiao-ping1,2, ZHANG Jun-li3, FU Jun-sheng1,2( )
)
Received:2021-04-07
Published:2021-11-26
Online:2021-12-03
摘要:
前期研究表明,蛹虫草活性成分虫草素能有效抑制乳腺癌移植瘤的生长和转移,但其作用机制未清楚。本研究取三阴性乳腺癌移植瘤样品,通过转录组测序技术分析虫草素抗乳腺癌移植瘤的分子作用机制。结果表明,以P<0.05、log2|FC|>1为标准,筛选获得584个差异表达基因,其中上调基因69个,下调基因515个;KEGG分析表明,虫草素对乳腺癌移植瘤的分子调控涉及Hippo signaling pathway、Hedgehog signaling pathway、ECM-receptor interction、TGF-beta signaling pathway等多条癌症相关信号通路;GO分析表明,这些差异基因富集在biological adhesion、growth、metabolic process、locomotion和biological process等16个生物学过程中,提示虫草素对乳腺癌移植瘤生长与转移有极显著的调节作用;进一步对生长和转移过程的基因进行PPI互作分析,提示Hedgehog signaling pathway、Hippo signaling pathway是虫草素抗三阴性乳腺癌的重要潜在作用通路。研究结果提示Hippo-Yap/TAZ信号通路和Hedgehog-Gli信号通路为潜在作用通路,为进一步理解虫草素治疗三阴性乳腺癌的分子机制奠定了基础。
黄海辰, 吴文雅, 戚梦, 薛帆正, 吴小平, 张君丽, 傅俊生. 虫草素抗三阴性乳腺癌的转录组学分析[J]. 生物技术通报, 2021, 37(11): 72-80.
HUANG Hai-chen, WU Wen-ya, QI Meng, XUE Fan-zheng, WU Xiao-ping, ZHANG Jun-li, FU Jun-sheng. RNA-Seq Analysis of Cordycepin Against Triple-negative Breast Cancer[J]. Biotechnology Bulletin, 2021, 37(11): 72-80.
| 基因Gene | 引物序列Primer sequence(5'-3') |
|---|---|
| PDK4 | F:5'-GAGGATTACTGACCGCCTCTTTAG-3' R:5'-TTCCGGGAATTGTCCATCAC-3' |
| CD24 | F:5'-CCCACGCAGATTTATTCCAG-3' R:5'-GACTTCCAGACGCCATTTG-3' |
| MMP7 | F:5'-AGCCAAACTCAAGGAGATGC-3' R:5'-GCCAATCATGATGTCAGCAG-3' |
| SHH | F:5'-AGGGCACCATTCTCATCAAC-3' R:5'-GGAGCGGTTAGGGCTACTCT-3' |
| GLI2 | F:5'-GCCCTT CCTGAAAAGAAGAC-3' R:5'-CATTGGAGA AACAGGATTGG-3' |
| GADPH | F:5'-GAAGGACTCATGACCACAG-3' R:5'-CTTCACCACCTTCTTGATG-3' |
表1 实时荧光定量qPCR的引物序列和片段长度
Table 1 Primers sequences and fragment lengths for qPCR
| 基因Gene | 引物序列Primer sequence(5'-3') |
|---|---|
| PDK4 | F:5'-GAGGATTACTGACCGCCTCTTTAG-3' R:5'-TTCCGGGAATTGTCCATCAC-3' |
| CD24 | F:5'-CCCACGCAGATTTATTCCAG-3' R:5'-GACTTCCAGACGCCATTTG-3' |
| MMP7 | F:5'-AGCCAAACTCAAGGAGATGC-3' R:5'-GCCAATCATGATGTCAGCAG-3' |
| SHH | F:5'-AGGGCACCATTCTCATCAAC-3' R:5'-GGAGCGGTTAGGGCTACTCT-3' |
| GLI2 | F:5'-GCCCTT CCTGAAAAGAAGAC-3' R:5'-CATTGGAGA AACAGGATTGG-3' |
| GADPH | F:5'-GAAGGACTCATGACCACAG-3' R:5'-CTTCACCACCTTCTTGATG-3' |
| 样品 Sample | 浓度 Concentration/(ng·μL-1) | 体积 Volume /μL | 总量 Total /μg | RIN值 RIN value | 文库类型 Library type | 结论 Conclusion |
|---|---|---|---|---|---|---|
| C0R1 | 295 | 35 | 10.33 | 6.8 | RNAseq | A |
| C0R2 | 528 | 35 | 18.46 | 7.9 | RNAseq | A |
| C0R3 | 761 | 35 | 26.67 | 9.3 | RNAseq | A |
| C0N1 | 652 | 76 | 49.55 | 8.3 | RNAseq | A |
| C0N2 | 695 | 76 | 52.78 | 9.3 | RNAseq | A |
| C0N3 | 602 | 74 | 44.51 | 9.3 | RNAseq | A |
表2 RNA测序质量评估
Table 2 RNA sequencing quality assessment
| 样品 Sample | 浓度 Concentration/(ng·μL-1) | 体积 Volume /μL | 总量 Total /μg | RIN值 RIN value | 文库类型 Library type | 结论 Conclusion |
|---|---|---|---|---|---|---|
| C0R1 | 295 | 35 | 10.33 | 6.8 | RNAseq | A |
| C0R2 | 528 | 35 | 18.46 | 7.9 | RNAseq | A |
| C0R3 | 761 | 35 | 26.67 | 9.3 | RNAseq | A |
| C0N1 | 652 | 76 | 49.55 | 8.3 | RNAseq | A |
| C0N2 | 695 | 76 | 52.78 | 9.3 | RNAseq | A |
| C0N3 | 602 | 74 | 44.51 | 9.3 | RNAseq | A |

图2 样品reads分类图 CON1:对照组1;CON2:对照组2;CON3:对照组3;COR1:实验组1;COR2:实验组2;COR3:实验组3;Adaptor:含有接头序列的reads数占总reads数的比例;High rate:含未知碱基的reads数占reads总数的比例;Low quality:从原始序列数据中去除杂质获得的数据与总读数之比;High quality:总读数
Fig.2 Classification chart for sample reads CON1:Control group 1. CON2:Control group 2. CON3:Control group 3. COR1:Cordycepin 1. COR2:Cordycepin 2. COR3:Cordycepin 3. Adaptor:The ratio of the number of reads containing the adaptor sequence to the total number of reads. High rate:The ratio of the number of reads containing unknown bases to the total number of reads. Low quality:The ratio of the data obtained by removing impurities from the original sequence data to the total number of reads. High quality:Total readings
| 样品名 Sample | 总reads数量 Number of total reads | 比对上rRNA的 reads 数及占总数的比例 Mapped reads(ratio in total) | 未比对上rRNA的 reads 数及占总数的比例 Unmapped reads(ratio in total) |
|---|---|---|---|
| CON1 | 53195320 | 410412(0.77%) | 52784908(99.23%) |
| CON2 | 69510386 | 493598(0.71%) | 69016788(99.29%) |
| CON3 | 65958874 | 550350(0.83%) | 65408524(99.17%) |
| COR1 | 90497008 | 812122(0.90%) | 89684886(99.10%) |
| COR2 | 78992736 | 387314(0.49%) | 78605422(99.51%) |
| COR3 | 56738684 | 310270(0.55%) | 56428414(99.45%) |
表3 HQ clean data与rRNA的比对统计表
Table 3 HQ clean data and rRNA alignment statistics
| 样品名 Sample | 总reads数量 Number of total reads | 比对上rRNA的 reads 数及占总数的比例 Mapped reads(ratio in total) | 未比对上rRNA的 reads 数及占总数的比例 Unmapped reads(ratio in total) |
|---|---|---|---|
| CON1 | 53195320 | 410412(0.77%) | 52784908(99.23%) |
| CON2 | 69510386 | 493598(0.71%) | 69016788(99.29%) |
| CON3 | 65958874 | 550350(0.83%) | 65408524(99.17%) |
| COR1 | 90497008 | 812122(0.90%) | 89684886(99.10%) |
| COR2 | 78992736 | 387314(0.49%) | 78605422(99.51%) |
| COR3 | 56738684 | 310270(0.55%) | 56428414(99.45%) |
| 样品名 Sample | 总reads数量 Total reads | 未比对上参考基因组的 reads 数及占总数的比例 Unmapped reads(ratio in total) | 唯一比对上参考基因组的 reads 数及占总数的比例 Unique mapped reads(ratio in total) | 多处比对上参考基因组的 reads 数及占总数的比例 Multiple mapped reads(ratio in total) | 比对上参考基因组的 reads占总数的比例 Mapping ratio |
|---|---|---|---|---|---|
| CON1 | 52784908 | 19192339(36.36%) | 33372603(63.22%) | 219966(0.42%) | 63.64% |
| CON2 | 69016788 | 24438545(35.41%) | 44287405(64.17%) | 290838(0.42%) | 64.59% |
| CON3 | 65408524 | 14470073(22.12%) | 50589691(77.34%) | 348760(0.53%) | 77.88% |
| COR1 | 89684886 | 16369614(18.25%) | 72826044(81.20%) | 489228(0.55%) | 81.75% |
| COR2 | 78605422 | 19831015(25.23%) | 58342213(74.22%) | 432194(0.55%) | 74.77% |
| COR3 | 56428414 | 18462152(32.72%) | 37706418(66.82%) | 259844(0.46%) | 67.28% |
表4 比对核糖体后得到的unmapped reads与参考基因组的比对统计表
Table 4 Comparison of unmapped reads and reference genomes obtained after ribosomal alignment
| 样品名 Sample | 总reads数量 Total reads | 未比对上参考基因组的 reads 数及占总数的比例 Unmapped reads(ratio in total) | 唯一比对上参考基因组的 reads 数及占总数的比例 Unique mapped reads(ratio in total) | 多处比对上参考基因组的 reads 数及占总数的比例 Multiple mapped reads(ratio in total) | 比对上参考基因组的 reads占总数的比例 Mapping ratio |
|---|---|---|---|---|---|
| CON1 | 52784908 | 19192339(36.36%) | 33372603(63.22%) | 219966(0.42%) | 63.64% |
| CON2 | 69016788 | 24438545(35.41%) | 44287405(64.17%) | 290838(0.42%) | 64.59% |
| CON3 | 65408524 | 14470073(22.12%) | 50589691(77.34%) | 348760(0.53%) | 77.88% |
| COR1 | 89684886 | 16369614(18.25%) | 72826044(81.20%) | 489228(0.55%) | 81.75% |
| COR2 | 78605422 | 19831015(25.23%) | 58342213(74.22%) | 432194(0.55%) | 74.77% |
| COR3 | 56428414 | 18462152(32.72%) | 37706418(66.82%) | 259844(0.46%) | 67.28% |
| 各分组名称 Group name | 检测到的已知基因数及比率 Number of known genes(ratio) | 检测到的新基因数目 Number of newly detected genes | 所有基因数目 Number of all genes |
|---|---|---|---|
| COR | 15 730(80.23%) | 1 405 | 17 135 |
| CON | 15 658(79.86%) | 1 394 | 17 052 |
表5 各组检测基因数目统计
Table 5 Statistics of the number of detected genes in each group
| 各分组名称 Group name | 检测到的已知基因数及比率 Number of known genes(ratio) | 检测到的新基因数目 Number of newly detected genes | 所有基因数目 Number of all genes |
|---|---|---|---|
| COR | 15 730(80.23%) | 1 405 | 17 135 |
| CON | 15 658(79.86%) | 1 394 | 17 052 |
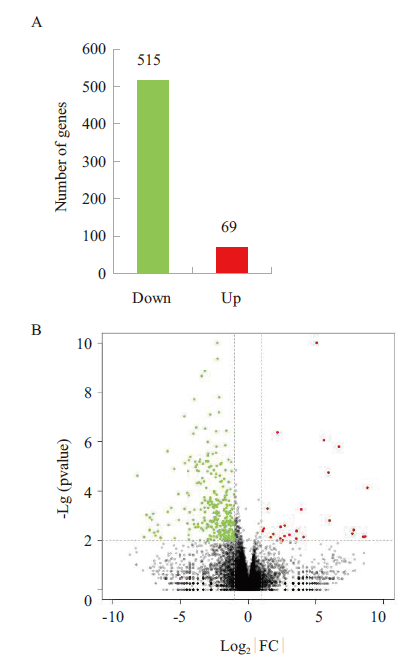
图3 分组间差异统计 A:样品间差异基因统计柱状图;B:CON vs COR差异基因火山图
Fig.3 Inter-group difference statistics A:Inter-sample differential gene statistics histogram. B:CON vs COR differential gene volcano map
| 检测项目 Project | MMP7 | PDK4 | CD24 | LIFR | GLI2 |
|---|---|---|---|---|---|
| RNA-seq | -2.55 | -2.09 | -2.37 | -1.35 | -2.34 |
| qRT-PCR/GAPDH | -2.05 | -1.25 | -2.31 | -2.07 | -2.87 |
表6 5个随机基因的RNA-seq与qPCR表达量对比
Table 6 Comparison between RNA-seq and qPCR expression levels of 5 random genes
| 检测项目 Project | MMP7 | PDK4 | CD24 | LIFR | GLI2 |
|---|---|---|---|---|---|
| RNA-seq | -2.55 | -2.09 | -2.37 | -1.35 | -2.34 |
| qRT-PCR/GAPDH | -2.05 | -1.25 | -2.31 | -2.07 | -2.87 |
| 条目 Term | 基因数量 Number of genes | 基因占比 Percentage/% | 基因 Gene |
|---|---|---|---|
| 生长 Growth | 21 | 2.142 | GLI2, ULK2, HNF4A, COBL, SEMA5A, WNT5A, IGFBP2, IGFBP5, PROX1, STRA6, DUOX2, NKD1, NLGN4X, DRAXIN, SHH, ISLR2, KLK6, HOPX, PSAPL1, SOX2, SELENOP |
| 黏附 Biological adhesion | 31 | 2.683 | ADAM28, GLI2, HNF4A, LHB, MDFI, CLIC5, WNT5A, IGFBP2, IGFBP5, CNR1, NDP, HOXD10, HAVCR2, NKX21, MMP7, STRA6, LGR5, DUOX2, ACE, DNALI1, HPGD, SHH, SPINT1, PDE3A, ADGRG2, PSAPL1, CADM1, TMEM119, CCIN, PAX5, SELENOP |
| 运动 Locomotion | 28 | 1.374 | GLI2, TESC, HNF4A, FLT1, FA2H, SEMA5A, WNT5A, IGFBP2, IGFBP5, PROX1, NDP, HAVCR2, LGR5, VASH2, CD109, ACE, TFF1, SHH, CTHRC1, SPINT1, ATOH8, TP53I11, SOX2, TMEM119, GNG2, TNFRSF18, SULF2, CD24 |
表7 生长与转移差异基因
Table 7 Growth and metastasis difference gene
| 条目 Term | 基因数量 Number of genes | 基因占比 Percentage/% | 基因 Gene |
|---|---|---|---|
| 生长 Growth | 21 | 2.142 | GLI2, ULK2, HNF4A, COBL, SEMA5A, WNT5A, IGFBP2, IGFBP5, PROX1, STRA6, DUOX2, NKD1, NLGN4X, DRAXIN, SHH, ISLR2, KLK6, HOPX, PSAPL1, SOX2, SELENOP |
| 黏附 Biological adhesion | 31 | 2.683 | ADAM28, GLI2, HNF4A, LHB, MDFI, CLIC5, WNT5A, IGFBP2, IGFBP5, CNR1, NDP, HOXD10, HAVCR2, NKX21, MMP7, STRA6, LGR5, DUOX2, ACE, DNALI1, HPGD, SHH, SPINT1, PDE3A, ADGRG2, PSAPL1, CADM1, TMEM119, CCIN, PAX5, SELENOP |
| 运动 Locomotion | 28 | 1.374 | GLI2, TESC, HNF4A, FLT1, FA2H, SEMA5A, WNT5A, IGFBP2, IGFBP5, PROX1, NDP, HAVCR2, LGR5, VASH2, CD109, ACE, TFF1, SHH, CTHRC1, SPINT1, ATOH8, TP53I11, SOX2, TMEM119, GNG2, TNFRSF18, SULF2, CD24 |
| 序号 Index | 名称 Name | P值 P-value | 调整后P值 Adjusted P-value | 综合得分 Combined score |
|---|---|---|---|---|
| 1 | Hedgehog signaling pathway | 0.0006293 | 0.02289 | 13.42 |
| 2 | Basal cell carcinoma | 0.0008323 | 0.02289 | 13.28 |
| 3 | Hippo signaling pathway | 0.001736 | 0.03183 | 10.73 |
| 4 | Focal adhesion | 0.004714 | 0.05655 | 9.89 |
| 5 | T cell receptor signaling pathway | 0.005141 | 0.05655 | 9.33 |
| 6 | Pathways in cancer | 0.01061 | 0.09472 | 8.98 |
| 7 | Cell adhesion molecules(CAMs) | 0.01206 | 0.09472 | 6.99 |
| 8 | Fc epsilon RI signaling pathway | 0.02185 | 0.1247 | 6.78 |
表8 基于Enrichr信号通路富集分析
Table 8 Enichr-based signal path enrichment analysis
| 序号 Index | 名称 Name | P值 P-value | 调整后P值 Adjusted P-value | 综合得分 Combined score |
|---|---|---|---|---|
| 1 | Hedgehog signaling pathway | 0.0006293 | 0.02289 | 13.42 |
| 2 | Basal cell carcinoma | 0.0008323 | 0.02289 | 13.28 |
| 3 | Hippo signaling pathway | 0.001736 | 0.03183 | 10.73 |
| 4 | Focal adhesion | 0.004714 | 0.05655 | 9.89 |
| 5 | T cell receptor signaling pathway | 0.005141 | 0.05655 | 9.33 |
| 6 | Pathways in cancer | 0.01061 | 0.09472 | 8.98 |
| 7 | Cell adhesion molecules(CAMs) | 0.01206 | 0.09472 | 6.99 |
| 8 | Fc epsilon RI signaling pathway | 0.02185 | 0.1247 | 6.78 |
| [1] | 袁晓玲, 尤婧蓉, 沈莉莉, 等. 乳腺癌化学治疗患者更年期症状相关生活质量及影响因素研究[J]. 上海交通大学学报:医学版, 2018, 38(12): 1457-1462. |
| Yuan XL, You JR, Shen LL, et al. Study on the predictors of the menopausal symptoms related quality of life in patients with breast cancer undergoing chemotherapy[J]. J Shanghai Jiao Tong Univ:Med Sci, 2018, 38(12): 1457-1462. | |
| [2] | 赵易, 赵庆丽, 马骥. 二甲双胍联合多西他赛对乳腺癌细胞MDA-MB-231增殖和凋亡的影响[J]. 现代肿瘤医学, 2018, 26(1): 10-13. |
| Zhao Y, Zhao QL, Ma J. Proliferation and apoptosis effect of combining metformin and docetaxel on breast cancer cells[J]. J Mod Oncol, 2018, 26(1): 10-13. | |
| [3] | 倪晨, 李婷, 吴振华, 等. 三阴性乳腺癌化疗进展[J]. 中国癌症杂志, 2014, 24(4): 316-320. |
| Ni C, Li T, Wu ZH, et al. Progress on chemotherapy for triple negative breast cancer[J]. China Oncol, 2014, 24(4): 316-320. | |
| [4] |
Auvinen P, Tammi R, Tammi M, et al. Expression of CD44s, CD44v3 and CD44v6 in benign and malignant breast lesions:correlation and colocalization with hyaluronan[J]. Histopathology, 2005, 47(4): 420-428.
pmid: 16178897 |
| [5] |
Liu C, Qi M, Li L, et al. Natural cordycepin induces apoptosis and suppresses metastasis in breast cancer cells by inhibiting the Hedgehog pathway[J]. Food Funct, 2020, 11(3): 2107-2116.
doi: 10.1039/C9FO02879J URL |
| [6] |
Habib JG, O’Shaughnessy JA. The hedgehog pathway in triple-negative breast cancer[J]. Cancer Med, 2016, 5(10): 2989-3006.
doi: 10.1002/cam4.2016.5.issue-10 URL |
| [7] |
Pandolfi S, Stecca B. Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways:implications for cancer therapy[J]. Expert Rev Mol Med, 2015, 17: e5.
doi: 10.1017/erm.2015.3 URL |
| [8] |
Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease[J]. Nat Rev Mol Cell Biol, 2013, 14(7): 416-429.
doi: 10.1038/nrm3598 URL |
| [9] |
Tang JY, Mackay-Wiggan JM, Aszterbaum M, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome[J]. N Engl J Med, 2012, 366(23): 2180-2188.
doi: 10.1056/NEJMoa1113538 URL |
| [10] |
Kayed H, Kleeff J, Osman T, et al. Hedgehog signaling in the normal and diseased pancreas[J]. Pancreas, 2006, 32(2): 119-129.
doi: 10.1097/01.mpa.0000202937.55460.0c URL |
| [11] | Park JH, Shin JE, Park HW. The role of hippo pathway in cancer stem cell biology[J]. Mol Cells, 2018, 41(2): 83-92. |
| [12] |
Cordenonsi M, Zanconato F, Azzolin L, et al. The hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells[J]. Cell, 2011, 147(4): 759-772.
doi: 10.1016/j.cell.2011.09.048 URL |
| [13] |
Maugeri-Saccà M, De Maria R. Hippo pathway and breast cancer stem cells[J]. Crit Rev Oncol Hematol, 2016, 99: 115-122.
doi: 10.1016/j.critrevonc.2015.12.004 pmid: 26725175 |
| [14] |
Moroishi T, Hayashi T, Pan WW, et al. The hippo pathway kinases LATS1/2 suppress cancer immunity[J]. Cell, 2016, 167(6): 1525-1539.e17.
doi: S0092-8674(16)31526-4 pmid: 27912060 |
| [15] |
Britschgi A, Duss S, Kim S, et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα[J]. Nature, 2017, 541(7638): 541-545.
doi: 10.1038/nature20829 URL |
| [16] |
Ercolani C, Di Benedetto A, Terrenato I, et al. Expression of phosphorylated Hippo pathway kinases(MST1/2 and LATS1/2)in HER2-positive and triple-negative breast cancer patients treated with neoadjuvant therapy[J]. Cancer Biol Ther, 2017, 18(5): 339-346.
doi: 10.1080/15384047.2017.1312230 URL |
| [17] |
Heidary Arash E, Shiban A, Song S, et al. MARK4 inhibits Hippo signaling to promote proliferation and migration of breast cancer cells[J]. EMBO Rep, 2017, 18(3): 420-436.
doi: 10.15252/embr.201642455 pmid: 28183853 |
| [18] | Yao CB, Zhou X, Chen CS, et al. The regulatory mechanisms and functional roles of the Hippo signaling pathway in breast cancer[J]. Yi Chuan, 2017, 39(7): 617-629. |
| [19] |
Sulaiman A, McGarry S, Li L, et al. Dual inhibition of Wnt and Yes-associated protein signaling retards the growth of triple-negative breast cancer in both mesenchymal and epithelial states[J]. Mol Oncol, 2018, 12(4): 423-440.
doi: 10.1002/mol2.2018.12.issue-4 URL |
| [20] |
Andrade D, Mehta M, Griffith J, et al. YAP1 inhibition radiosensitizes triple negative breast cancer cells by targeting the DNA damage response and cell survival pathways[J]. Oncotarget, 2017, 8(58): 98495-98508.
doi: 10.18632/oncotarget.21913 pmid: 29228705 |
| [21] |
Cotton JL, Li Q, Ma L, et al. YAP/TAZ and hedgehog coordinate growth and patterning in gastrointestinal mesenchyme[J]. Dev Cell, 2017, 43(1): 35-47.e4.
doi: 10.1016/j.devcel.2017.08.019 URL |
| [22] |
Watanabe Y, Miyasaka KY, Kubo A, et al. Notch and Hippo signaling converge on Strawberry Notch 1(Sbno1)to synergistically activate Cdx2 during specification of the trophectoderm[J]. Sci Rep, 2017, 7(1): 1-17.
doi: 10.1038/s41598-016-0028-x URL |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [3] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [4] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [5] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [6] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [7] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [8] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [9] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [10] | 徐俊, 叶雨晴, 牛雅静, 黄河, 张蒙蒙. 菊花根状茎发育的转录组分析[J]. 生物技术通报, 2023, 39(10): 231-245. |
| [11] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [12] | 辛建攀, 李燕, 赵楚, 田如男. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210. |
| [13] | 许瑾, 李涛, 李楚琳, 朱顺妮, 王忠铭, 向文洲. 温度对真眼点藻生长、总脂及二十碳五烯酸(EPA)合成的影响[J]. 生物技术通报, 2022, 38(6): 261-271. |
| [14] | 熊和丽, 沙茜, 刘韶娜, 相德才, 张斌, 赵智勇. 单细胞转录组测序技术在动物上的应用研究[J]. 生物技术通报, 2022, 38(3): 226-233. |
| [15] | 张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||