








生物技术通报 ›› 2021, Vol. 37 ›› Issue (8): 1-11.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0861
• 代谢生物学专题 • 下一篇
收稿日期:2021-07-08
出版日期:2021-08-26
发布日期:2021-09-10
作者简介:张凤,女,博士,研究方向:作物代谢组学;E-mail: 基金资助:Received:2021-07-08
Published:2021-08-26
Online:2021-09-10
摘要:
近年随着持续而又复杂环境的改变,自然界中生物和非生物胁迫频繁爆发,多种逆境胁迫严重影响了植物的正常生长和发育,尤其是农作物产量。逆境胁迫下植物体内代谢物的重塑是其基因与环境因素共同作用的结果,是植物体生理表型与体内生化水平的直接体现,逆境胁迫下代谢组的重塑很大程度上反映了植物体对逆境胁迫的响应和防御。代谢组学的兴起,为研究植物体内不同组织及其在不同逆境胁迫下代谢物的重塑提供了可靠的研究手段,同时代谢组与基因组、转录组、蛋白组以及表型组的整合,尤其是代谢组与基因组整合形成的代谢组-基因组关联分析在揭示植物响应及适应逆境胁迫的遗传基础、提高农作物产量以及培育耐受逆境胁迫品种等方面具有重要作用。本文综述了逆境胁迫下植物代谢组学的研究方法、逆境胁迫下植物代谢组重塑的多样性以及逆境胁迫下植物代谢组的遗传基础研究进展,并展望了应用代谢组学研究植物逆境生物学的应用前景和局限性。
张凤, 陈伟. 代谢组学在植物逆境生物学中的研究进展[J]. 生物技术通报, 2021, 37(8): 1-11.
ZHANG Feng, CHEN Wei. Research Progress of Metabolomics in Plant Stress Biology[J]. Biotechnology Bulletin, 2021, 37(8): 1-11.
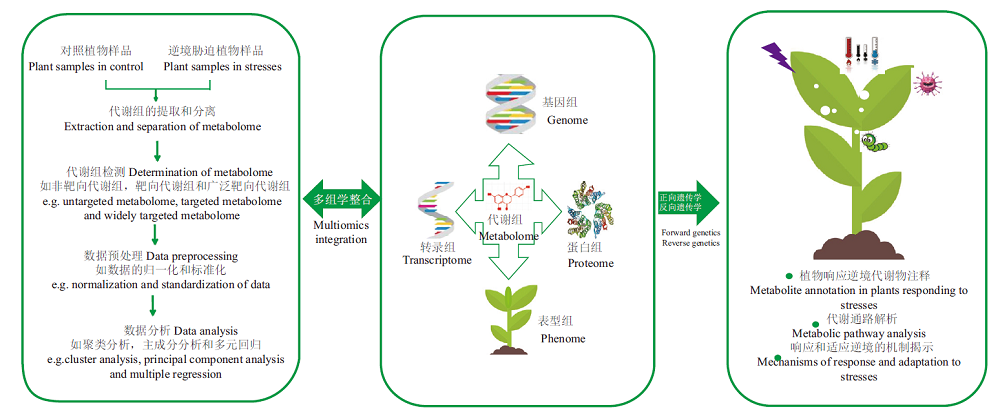
图1 逆境胁迫下植物代谢组学的研究流程 逆境胁迫下植物代谢组学的研究流程包括逆境胁迫与对照条件下植物样品的制备、代谢物的提取、代谢物的检测、数据采集、数据预处理以及数据的初步分析等。为了进一步分析逆境胁迫下代谢组,代谢组与基因组、转录组、蛋白组以及表型组等多组学整合,结合反向遗传学研究方法,对植物响应逆境的代谢物注释、代谢通路解析以及解析植物响应和适应逆境的调控机制
Fig.1 Research process of plant metabolomics under stresses The research process of plant metabolomics under stresses includes the preparation of plant samples under stresses and control conditions,the extraction of metabolites,the detection of metabolites,data collection,data preprocessing,and the data preliminary analysis. In order to further explore the metabolome data under stresses,the integration of metabolome with genome,transcriptome,proteome and phenome,and with reverse genetic research methods,can annotate the metabolites,analyze the metabolic pathways and explain the regulatory mechanism of plant response and adaptation to stresses
| 代谢物名称 Metabolite name | 代谢物功能 Metabolite function | 代谢物类别 Metabolite category | 物种 Species | 参考文献 References |
|---|---|---|---|---|
| 尼酸、羟基肉桂酸和木质素 | 增强稻瘟病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 茉莉酸、二氢甘氨酸、山奈酚和甲氧基肉桂酸 | 增强赤霉病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 黄烷醇、香豆素和异黄酮 | 增强赤霉病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 脂肪酸、可溶性糖、苯甲醛、黄酮醇和葡萄素 | 增强霜霉病抗性 | 初生和次生代谢物 | 葡萄Vitis vinifera | [ |
| 脯氨酸、甜菜苷和槲皮素 | 增强干旱胁迫抗性 | 初生和次生代谢物 | 豇豆Vigna unguiculata | [ |
| 脯氨酸、组氨酸、异亮氨酸和色氨酸 | 增强干旱胁迫抗性 | 初生代谢物 | 鹰嘴豆Cicer arietinum | [ |
| 甜菜碱、脯氨酸、多胺以和羟基醇 | 增强盐胁迫抗性 | 初生和次生代谢物 | 高粱Sorghum bicolor | [ |
| 丝氨酸、山梨糖、果糖和戊酸 | 增强盐胁迫抗性 | 初生代谢物 | 番茄Solanum lycopersicum | [ |
| 脯氨酸、戊二酸、半乳糖酸和抗坏血酸五羟色胺和褪黑素 | 增强盐胁迫抗性 增强冷和冻胁迫抗性 | 初生代谢物 次生代谢物 | 大豆Glycine max 番茄Solanum lycopersicum | [ [ |
| 褪黑素 | 增强冷胁迫抗性 | 次生代谢物 | 水稻Oryza sativa | [ |
| 槲皮素、山奈酚和矢车菊素 | 增强氧化和干旱胁迫抗性 | 次生代谢物 | 拟南芥Arabidopsis thaliana | [ |
| 黄酮醇 | 增强紫外线胁迫抗性 | 次生代谢物 | 拟南芥Arabidopsis thaliana | [ |
| 2-己醛和3-己醛 | 增强洪涝胁迫抗性 | 初生代谢物 | 葡萄Vitis vinifera | [ |
| 葡萄糖、棉子糖、果糖、脯氨酸和色氨酸 | 增强寒胁迫抗性 | 初生代谢物 | 水稻Oryza sativa | [ |
| 硫胺、生育酚、脯氨酸、丙氨酸和氨基丁酸 | 增强纹枯病抗性 | 初生代谢物 | 大豆Glycine max | [ |
| 乙烯和茉莉酸 | 增强干旱胁迫抗性 | 初生代谢物 | 番茄Solanum lycopersicum | [ |
| 磷脂酸、羟基肉桂酸和槲皮素 | 增强赤霉病抗性 | 初生和次生代谢物 | 小麦Triticum aestivum | [ |
| 苯醌、金雀异黄酮和毛地黄黄酮 | 增强尖孢镰刀菌抗性 | 初生和次生代谢物 | 鹰嘴豆Cicer arietinum | [ |
| 脯氨酸、香豆酸和绿原酸 | 增强干旱胁迫抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 草酸 | 增强小麦黑穗病抗性 | 初生代谢物 | 小麦Triticum aestivum | [ |
| γ-生育酚、谷胱甘肽和琥珀酸 | 增强干旱及热胁迫抗性 | 初生代谢物 | 大麦Hordeum vulgare | [ |
| 肉桂酸和木质素 | 增强叶枯病和灰斑病抗性 | 次生代谢物 | 玉米Zea mays | [ |
| 古龙糖、抗坏血酸、葡萄糖酸和苏氨酸 | 增强干旱胁迫抗性 | 初生代谢物 | 小麦Triticum aestivum | [ |
| 山奈酚、毛地黄黄酮和麦黄酮木脂素 | 增强紫外线胁迫抗性 | 次生代谢物 | 水稻Oryza sativa | [ |
表1 植物响应逆境胁迫代谢物研究列表
Table1 Research list of plant metabolites in response to stresses
| 代谢物名称 Metabolite name | 代谢物功能 Metabolite function | 代谢物类别 Metabolite category | 物种 Species | 参考文献 References |
|---|---|---|---|---|
| 尼酸、羟基肉桂酸和木质素 | 增强稻瘟病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 茉莉酸、二氢甘氨酸、山奈酚和甲氧基肉桂酸 | 增强赤霉病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 黄烷醇、香豆素和异黄酮 | 增强赤霉病抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 脂肪酸、可溶性糖、苯甲醛、黄酮醇和葡萄素 | 增强霜霉病抗性 | 初生和次生代谢物 | 葡萄Vitis vinifera | [ |
| 脯氨酸、甜菜苷和槲皮素 | 增强干旱胁迫抗性 | 初生和次生代谢物 | 豇豆Vigna unguiculata | [ |
| 脯氨酸、组氨酸、异亮氨酸和色氨酸 | 增强干旱胁迫抗性 | 初生代谢物 | 鹰嘴豆Cicer arietinum | [ |
| 甜菜碱、脯氨酸、多胺以和羟基醇 | 增强盐胁迫抗性 | 初生和次生代谢物 | 高粱Sorghum bicolor | [ |
| 丝氨酸、山梨糖、果糖和戊酸 | 增强盐胁迫抗性 | 初生代谢物 | 番茄Solanum lycopersicum | [ |
| 脯氨酸、戊二酸、半乳糖酸和抗坏血酸五羟色胺和褪黑素 | 增强盐胁迫抗性 增强冷和冻胁迫抗性 | 初生代谢物 次生代谢物 | 大豆Glycine max 番茄Solanum lycopersicum | [ [ |
| 褪黑素 | 增强冷胁迫抗性 | 次生代谢物 | 水稻Oryza sativa | [ |
| 槲皮素、山奈酚和矢车菊素 | 增强氧化和干旱胁迫抗性 | 次生代谢物 | 拟南芥Arabidopsis thaliana | [ |
| 黄酮醇 | 增强紫外线胁迫抗性 | 次生代谢物 | 拟南芥Arabidopsis thaliana | [ |
| 2-己醛和3-己醛 | 增强洪涝胁迫抗性 | 初生代谢物 | 葡萄Vitis vinifera | [ |
| 葡萄糖、棉子糖、果糖、脯氨酸和色氨酸 | 增强寒胁迫抗性 | 初生代谢物 | 水稻Oryza sativa | [ |
| 硫胺、生育酚、脯氨酸、丙氨酸和氨基丁酸 | 增强纹枯病抗性 | 初生代谢物 | 大豆Glycine max | [ |
| 乙烯和茉莉酸 | 增强干旱胁迫抗性 | 初生代谢物 | 番茄Solanum lycopersicum | [ |
| 磷脂酸、羟基肉桂酸和槲皮素 | 增强赤霉病抗性 | 初生和次生代谢物 | 小麦Triticum aestivum | [ |
| 苯醌、金雀异黄酮和毛地黄黄酮 | 增强尖孢镰刀菌抗性 | 初生和次生代谢物 | 鹰嘴豆Cicer arietinum | [ |
| 脯氨酸、香豆酸和绿原酸 | 增强干旱胁迫抗性 | 初生和次生代谢物 | 大麦Hordeum vulgare | [ |
| 草酸 | 增强小麦黑穗病抗性 | 初生代谢物 | 小麦Triticum aestivum | [ |
| γ-生育酚、谷胱甘肽和琥珀酸 | 增强干旱及热胁迫抗性 | 初生代谢物 | 大麦Hordeum vulgare | [ |
| 肉桂酸和木质素 | 增强叶枯病和灰斑病抗性 | 次生代谢物 | 玉米Zea mays | [ |
| 古龙糖、抗坏血酸、葡萄糖酸和苏氨酸 | 增强干旱胁迫抗性 | 初生代谢物 | 小麦Triticum aestivum | [ |
| 山奈酚、毛地黄黄酮和麦黄酮木脂素 | 增强紫外线胁迫抗性 | 次生代谢物 | 水稻Oryza sativa | [ |

图2 植物次生代谢响应逆境胁迫的调控网络 当植物体受到生物和非生物胁迫侵害时,体内响应蛋白(receptors)首先被激活。然后,响应蛋白激活下游的信号蛋白如蛋白激酶(RLKs)、丝裂原活化蛋白激酶(MAPKs)、转录因子(MYBs、WRKYs以及bZIPs等)以及热激蛋白(HSFs)等。最后,这些信号蛋白激活代谢途径相关基因如参与类黄酮代谢相关基因CHSs、FLSs和UGTs等以及参与萜类合成的相关基因GPSs、FPSs、GGPSs以及TPSs等的表达,促进黄酮类和萜类等物质的积累增强植物体对逆境胁迫的耐受性
Fig.2 Regulatory networks of plant secondary metabolism in response to stresses When plants are invaded by biotic and abiotic stresses,the receptors are firstly activated. The response proteins then activate downstream signaling proteins such as protein kinases(RLKs),mitogen-activated protein kinases(MAPKs),transcription factors(MYBs,WRKYs,bZIPs,etc.)and heat shock proteins(HSFs)etc. Finally,these signal proteins activate the expressions of genes related to metabolic pathways,such as CHSs,FLSs and UGTs involved in flavonoid metabolism,and GPSs,FPSs,GGPSs and TPSs involved in terpenoid synthesis to promote the accumulation of flavonoids and terpenoids and ultimately to enhance the tolerance of plant stresses
| [1] |
Elliott J, Deryng D, Müller C, et al. Constraints and potentials of future irrigation water availability on agricultural production under climate change[J]. PNAS, 2014, 111(9):3239-3244.
doi: 10.1073/pnas.1222474110 pmid: 24344283 |
| [2] |
Rortais A, Arnold G, Dorne JL, et al. Risk assessment of pesticides and other stressors in bees:Principles, data gaps and perspectives from the European Food Safety Authority[J]. Sci Total Environ, 2017, 587/588:524-537.
doi: 10.1016/j.scitotenv.2016.09.127 URL |
| [3] |
Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production[J]. Nature, 2016, 529(7584):84-87.
doi: 10.1038/nature16467 URL |
| [4] |
Michaletti A, Naghavi MR, Toorchi M, et al. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat[J]. Sci Rep, 2018, 8(1):5710.
doi: 10.1038/s41598-018-24012-y pmid: 29632386 |
| [5] |
Rudd JJ, Kanyuka K, Hassani-Pak K, et al. Transcriptome and metabolite profiling of the infection cycle of zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition[J]. Plant Physiol, 2015, 167(3):1158-1185.
doi: 10.1104/pp.114.255927 URL |
| [6] |
Liu XJ, Locasale JW. Metabolomics:a primer[J]. Trends Biochem Sci, 2017, 42(4):274-284.
doi: 10.1016/j.tibs.2017.01.004 URL |
| [7] |
Hong J, Yang LT, Zhang DB, et al. Plant metabolomics:an indispensable system biology tool for plant science[J]. Int J Mol Sci, 2016, 17(6):767.
doi: 10.3390/ijms17060767 URL |
| [8] | Bowne J, Bacic A, Tester M, et al. Abiotic stress and metabolomics[M]// Annual Plant Reviews Volume 43. Oxford, UK:Wiley-Blackwell, 2011:61-85. |
| [9] |
Nicholson JK, Lindon JC, Holmes E. 'Metabonomics':understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data[J]. Xenobiotica, 1999, 29(11):1181-1189.
pmid: 10598751 |
| [10] |
Fiehn O, Kopka J, Dörmann P, et al. Metabolite profiling for plant functional genomics[J]. Nat Biotechnol, 2000, 18(11):1157-1161.
pmid: 11062433 |
| [11] |
Arbona V, Iglesias DJ, Talón M, et al. Plant phenotype demarcation using nontargeted LC-MS and GC-MS metabolite profiling[J]. J Agric Food Chem, 2009, 57(16):7338-7347.
doi: 10.1021/jf9009137 URL |
| [12] |
Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks[J]. Comp Funct Genomics, 2001, 2(3):155-168.
doi: 10.1002/cfg.82 URL |
| [13] |
Vizán P, Mazurek S, Cascante M. Robust metabolic adaptation underlying tumor progression[J]. Metabolomics, 2008, 4(1):1-12.
doi: 10.1007/s11306-007-0101-3 URL |
| [14] |
Castro-Moretti FR, Gentzel IN, MacKey D, et al. Metabolomics as an emerging tool for the study of plant-pathogen interactions[J]. Metabolites, 2020, 10(2):52.
doi: 10.3390/metabo10020052 URL |
| [15] |
Bai Y, Kissoudis C, Yan Z, et al. Plant behaviour under combined stress:tomato responses to combined salinity and pathogen stress[J]. Plant J, 2018, 93(4):781-793.
doi: 10.1111/tpj.2018.93.issue-4 URL |
| [16] |
Akpinar BA, Avsar B, Lucas SJ, et al. Plant abiotic stress signaling[J]. Plant Signal Behav, 2012, 7(11):1450-1455.
doi: 10.4161/psb.21894 pmid: 22990453 |
| [17] |
Kessler A, Kalske A. Plant secondary metabolite diversity and species interactions[J]. Annu Rev Ecol Evol Syst, 2018, 49(1):115-138.
doi: 10.1146/annurev-ecolsys-110617-062406 URL |
| [18] |
Fang C, Fernie AR, Luo J. Exploring the diversity of plant metabolism[J]. Trends Plant Sci, 2019, 24(1):83-98.
doi: 10.1016/j.tplants.2018.09.006 URL |
| [19] |
Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology[J]. Annu Rev Plant Biol, 2010, 61:463-489.
doi: 10.1146/annurev.arplant.043008.092035 URL |
| [20] |
Ribbenstedt A, Ziarrusta H, Benskin JP. Development, characterization and comparisons of targeted and non-targeted metabolomics methods[J]. PLoS One, 2018, 13(11):e0207082.
doi: 10.1371/journal.pone.0207082 URL |
| [21] |
Luo P, Yin P, Hua R, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma[J]. Hepatology, 2018, 67(2):662-675.
doi: 10.1002/hep.v67.2 URL |
| [22] |
Song EH, Kim HJ, Jeong J, et al. A(1)H HR-MAS NMR-based metabolomic study for metabolic characterization of rice grain from various Oryza sativa L. cultivars[J]. J Agric Food Chem, 2016, 64(15):3009-3016.
doi: 10.1021/acs.jafc.5b05667 URL |
| [23] |
Wang JH, Byun J, Pennathur S. Analytical approaches to metabolomics and applications to systems biology[J]. Semin Nephrol, 2010, 30(5):500-511.
doi: 10.1016/j.semnephrol.2010.07.007 URL |
| [24] |
Pott DM, Osorio S, Vallarino JG. From central to specialized metabolism:an overview of some secondary compounds derived from the primary metabolism for their role in conferring nutritional and organoleptic characteristics to fruit[J]. Front Plant Sci, 2019, 10:835.
doi: 10.3389/fpls.2019.00835 URL |
| [25] |
Yang L, Wen KS, Ruan X, et al. Response of plant secondary metabolites to environmental factors[J]. Molecules, 2018, 23(4):762.
doi: 10.3390/molecules23040762 URL |
| [26] |
Tenenboim H, Brotman Y. Omic relief for the biotically stressed:metabolomics of plant biotic interactions[J]. Trends Plant Sci, 2016, 21(9):781-791.
doi: S1360-1385(16)30026-7 pmid: 27185334 |
| [27] |
Sulpice R, McKeown PC. Moving toward a comprehensive map of central plant metabolism[J]. Annu Rev Plant Biol, 2015, 66:187-210.
doi: 10.1146/annurev-arplant-043014-114720 pmid: 25621519 |
| [28] |
Hamany Djande CY, Pretorius C, Tugizimana F, et al. Metabolomics:a tool for cultivar phenotyping and investigation of grain crops[J]. Agronomy, 2020, 10(6):831.
doi: 10.3390/agronomy10060831 URL |
| [29] |
Peng B, Li H, Peng XX. Functional metabolomics:from biomarker discovery to metabolome reprogramming[J]. Protein Cell, 2015, 6(9):628-637.
doi: 10.1007/s13238-015-0185-x pmid: 26135925 |
| [30] |
Cajka T, Vaclavikova M, Dzuman Z, et al. Rapid LC-MS-based metabolomics method to study the Fusarium infection of barley[J]. J Sep Science, 2014, 37(8):912-919.
doi: 10.1002/jssc.v37.8 URL |
| [31] |
Obata T, Fernie AR. The use of metabolomics to dissect plant responses to abiotic stresses[J]. Cell Mol Life Sci, 2012, 69(19):3225-3243.
doi: 10.1007/s00018-012-1091-5 URL |
| [32] |
Parker D, Beckmann M, Zubair H, et al. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea[J]. Plant J, 2009, 59(5):723-737.
doi: 10.1111/tpj.2009.59.issue-5 URL |
| [33] |
Kumaraswamy KG, Kushalappa AC, Choo TM, et al. Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against Fusarium head blight(Fusarium graminearum)[J]. J Chem Ecol, 2011, 37(8):846-856.
doi: 10.1007/s10886-011-9989-1 pmid: 21701847 |
| [34] | Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism[J]. Plant Cell, 1995:1085-1097. |
| [35] |
Chitarrini G, Soini E, Riccadonna S, et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety[J]. Front Plant Sci, 2017, 8:1524.
doi: 10.3389/fpls.2017.01524 pmid: 28928759 |
| [36] |
Szabados L, Savouré A. Proline:a multifunctional amino acid[J]. Trends Plant Sci, 2010, 15(2):89-97.
doi: 10.1016/j.tplants.2009.11.009 pmid: 20036181 |
| [37] |
Lv WT, Lin B, Zhang M, et al. Proline accumulation is inhibitory to Arabidopsis seedlings during heat stress[J]. Plant Physiol, 2011, 156(4):1921-1933.
doi: 10.1104/pp.111.175810 URL |
| [38] |
Yadav AK, Carroll AJ, Estavillo GM, et al. Wheat drought tolerance in the field is predicted by amino acid responses to glasshouse-imposed drought[J]. J Exp Bot, 2019, 70(18):4931-4948.
doi: 10.1093/jxb/erz224 pmid: 31189018 |
| [39] |
Bettaieb I, Zakhama N, Wannes WA, et al. Water deficit effects on Salvia officinalis fatty acids and essential oils composition[J]. Sci Hortic, 2009, 120(2):271-275.
doi: 10.1016/j.scienta.2008.10.016 URL |
| [40] |
Liu CC, Liu YG, Guo K, et al. Effect of drought on pigments, osmotic adjustment and antioxidant enzymes in six woody plant species in Karst habitats of southwestern China[J]. Environ Exp Bot, 2011, 71(2):174-183.
doi: 10.1016/j.envexpbot.2010.11.012 URL |
| [41] |
Goufo P, Moutinho-Pereira JM, Jorge TF, et al. Cowpea(Vigna unguiculata L. walp. )metabolomics:osmoprotection as a physiological strategy for drought stress resistance and improved yield[J]. Front Plant Sci, 2017, 8:586.
doi: 10.3389/fpls.2017.00586 URL |
| [42] |
Khan N, Bano A, Rahman MA, et al. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea(Cicer arietinum)metabolome following long-term drought stress[J]. Plant Cell Environ, 2019, 42(1):115-132.
doi: 10.1111/pce.v42.1 URL |
| [43] |
Deinlein U, Stephan AB, Horie T, et al. Plant salt-tolerance mechanisms[J]. Trends Plant Sci, 2014, 19(6):371-379.
doi: 10.1016/j.tplants.2014.02.001 pmid: 24630845 |
| [44] |
de Lacerda CF, Cambraia J, Oliva MA, et al. Solute accumulation and distribution during shoot and leaf development in two Sorghum genotypes under salt stress[J]. Environ Exp Bot, 2003, 49(2):107-120.
doi: 10.1016/S0098-8472(02)00064-3 URL |
| [45] | Ye T, Shi H, Wang Y, et al. Contrasting proteomic and metabolomic responses of bermudagrass to drought and salt stresses[J]. Front Plant Sci, 2016, 7:1694. |
| [46] |
Yang DS, Zhang J, Li MX, et al. Metabolomics analysis reveals the salt-tolerant mechanism in Glycine soja[J]. J Plant Growth Regul, 2017, 36(2):460-471.
doi: 10.1007/s00344-016-9654-6 URL |
| [47] |
Ding F, Liu B, Zhang SX. Exogenous melatonin ameliorates cold-induced damage in tomato plants[J]. Sci Hortic, 2017, 219:264-271.
doi: 10.1016/j.scienta.2017.03.029 URL |
| [48] |
Fan JB, Xie Y, Zhang ZC, et al. Melatonin:a multifunctional factor in plants[J]. Int J Mol Sci, 2018, 19(5):1528.
doi: 10.3390/ijms19051528 URL |
| [49] |
Nakabayashi R, Yonekura-Sakakibara K, Urano K, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids[J]. Plant J, 2014, 77(3):367-379.
doi: 10.1111/tpj.2014.77.issue-3 URL |
| [50] |
Tohge T, Wendenburg R, Ishihara H, et al. Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae[J]. Nat Commun, 2016, 7:12399.
doi: 10.1038/ncomms12399 URL |
| [51] |
Ju YL, Yue XF, Zhao XF, et al. Physiological, micro-morphological and metabolomic analysis of grapevine(Vitis vinifera L.)leaf of plants under water stress[J]. Plant Physiol Biochem, 2018, 130:501-510.
doi: 10.1016/j.plaphy.2018.07.036 URL |
| [52] |
Ma'Ayan A. Complex systems biology[J]. J R Soc Interface, 2017, 14(134):20170391.
doi: 10.1098/rsif.2017.0391 URL |
| [53] | Parida AK, Panda A, Rangani J. Metabolomics-guided elucidation of abiotic stress tolerance mechanisms in plants[M]// Plant Metabolites and Regulation Under Environmental Stress. Amsterdam:Elsevier, 2018:89-131. |
| [54] |
Wang WS, Zhao XQ, Li M, et al. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling[J]. J Exp Bot, 2016, 67(1):405-419.
doi: 10.1093/jxb/erv476 URL |
| [55] |
Copley TR, Aliferis KA, Kliebenstein DJ, et al. An integrated RNAseq-1H NMR metabolomics approach to understand soybean primary metabolism regulation in response to Rhizoctonia foliar blight disease[J]. BMC Plant Biol, 2017, 17(1):84.
doi: 10.1186/s12870-017-1020-8 pmid: 28449662 |
| [56] |
Egea I, Albaladejo I, Meco V, et al. The drought-tolerant Solanum pennellii regulates leaf water loss and induces genes involved in amino acid and ethylene/jasmonate metabolism under dehydration[J]. Sci Rep, 2018, 8(1):2791.
doi: 10.1038/s41598-018-21187-2 URL |
| [57] |
Dhokane D, Karre S, Kushalappa AC, et al. Integrated metabolo-transcriptomics reveals Fusarium head blight candidate resistance genes in wheat QTL-Fhb2[J]. PLoS One, 2016, 11(5):e0155851.
doi: 10.1371/journal.pone.0155851 URL |
| [58] | Nussbaumer T, Warth B, Sharma S, et al. Joint transcriptomic and metabolomic analyses reveal changes in the primary metabolism and imbalances in the subgenome orchestration in the bread wheat molecular response to Fusarium graminearum[J]. G3:Bethesda, 2015, 5(12):2579-2592. |
| [59] | Meena KK, Sorty AM, Bitla UM, et al. Abiotic stress responses and microbe-mediated mitigation in plants:the omics strategies[J]. Front Plant Sci, 2017, 8:172. |
| [60] | Vo KTX, Rahman MM, Rahman MM, et al. Proteomics and metabolomics studies on the biotic stress responses of rice:an update[J]. Rice:N Y, 2021, 14(1):30. |
| [61] |
Kumar Y, Zhang L, Panigrahi P, et al. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics[J]. Plant Biotechnol J, 2016, 14(7):1589-1603.
doi: 10.1111/pbi.2016.14.issue-7 URL |
| [62] |
Chmielewska K, Rodziewicz P, Swarcewicz B, et al. Analysis of drought-induced proteomic and metabolomic changes in barley(Hordeum vulgare L.)leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance[J]. Front Plant Sci, 2016, 7:1108.
doi: 10.3389/fpls.2016.01108 pmid: 27512399 |
| [63] |
Pandey V, Singh M, Pandey D, et al. Integrated proteomics, genomics, metabolomics approaches reveal oxalic acid as pathogenicity factor in Tilletia indica inciting Karnal bunt disease of wheat[J]. Sci Rep, 2018, 8(1):7826.
doi: 10.1038/s41598-018-26257-z URL |
| [64] |
Wentzell AM, Rowe HC, Hansen BG, et al. Linking metabolic QTLs with network and Cis-eQTLs controlling biosynthetic pathways[J]. PLoS Genet, 2007, 3(9):1687-1701.
pmid: 17941713 |
| [65] |
Huang X, Zhao Y, Wei X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm[J]. Nat Genet, 2011, 44(1):32-39.
doi: 10.1038/ng.1018 URL |
| [66] |
Villate A, San Nicolas M, Gallastegi M, et al. Review:Metabolomics as a prediction tool for plants performance under environmental stress[J]. Plant Sci, 2021, 303:110789.
doi: 10.1016/j.plantsci.2020.110789 pmid: 33487364 |
| [67] |
Fang C, Luo J. Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism[J]. Plant J, 2019, 97(1):91-100.
doi: 10.1111/tpj.14097 URL |
| [68] |
Lisec J, Steinfath M, Meyer RC, et al. Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations[J]. Plant J, 2009, 59(5):777-788.
doi: 10.1111/tpj.2009.59.issue-5 URL |
| [69] |
Schauer N, Semel Y, Roessner U, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement[J]. Nat Biotechnol, 2006, 24(4):447-454.
doi: 10.1038/nbt1192 URL |
| [70] |
Templer SE, Ammon A, Pscheidt D, et al. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense[J]. J Exp Bot, 2017, 68(7):1697-1713.
doi: 10.1093/jxb/erx038 pmid: 28338908 |
| [71] |
Yang Q, He Y, Kabahuma M, et al. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens[J]. Nat Genet, 2017, 49(9):1364-1372.
doi: 10.1038/ng.3919 pmid: 28740263 |
| [72] |
Hill CB, Taylor JD, Edwards J, et al. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait Loci in bread wheat grown in a water-limited environment[J]. Plant Physiol, 2013, 162(3):1266-1281.
doi: 10.1104/pp.113.217851 URL |
| [73] |
Chen W, Gao Y, Xie W, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism[J]. Nat Genet, 2014, 46(7):714-721.
doi: 10.1038/ng.3007 pmid: 24908251 |
| [74] |
Zhu G, Wang S, Huang Z, et al. Rewiring of the fruit metabolome in tomato breeding[J]. Cell, 2018, 172(1/2):249-261.
doi: 10.1016/j.cell.2017.12.019 URL |
| [75] |
Zhang F, Guo H, Huang J, et al. A UV-B-responsive glycosyltransferase, OsUGT706C2, modulates flavonoid metabolism in rice[J]. Sci China Life Sci, 2020, 63(7):1037-1052.
doi: 10.1007/s11427-019-1604-3 pmid: 32112268 |
| [76] |
Stitt M, Sonnewald U. Regulation of metabolism in transgenic plants[J]. Annu Rev Plant Physiol Plant Mol Biol, 1995, 46(1):341-368.
doi: 10.1146/annurev.pp.46.060195.002013 URL |
| [77] |
Fernie AR, Tohge T. The genetics of plant metabolism[J]. Annu Rev Genet, 2017, 51:287-310.
doi: 10.1146/annurev-genet-120116-024640 URL |
| [78] |
Bringaud F, Biran M, Millerioux Y, et al. Combining reverse genetics and nuclear magnetic resonance-based metabolomics unravels trypanosome-specific metabolic pathways[J]. Mol Microbiol, 2015, 96(5):917-926.
doi: 10.1111/mmi.2015.96.issue-5 URL |
| [79] |
Belhaj K, Chaparro-Garcia A, Kamoun S, et al. Plant genome editing made easy:targeted mutagenesis in model and crop plants using the CRISPR/Cas system[J]. Plant Methods, 2013, 9(1):39.
doi: 10.1186/1746-4811-9-39 URL |
| [80] |
Xing HL, Dong L, Wang ZP, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants[J]. BMC Plant Biol, 2014, 14:327.
doi: 10.1186/s12870-014-0327-y URL |
| [1] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| [2] | 韩华蕊, 杨宇琭, 门艺涵, 韩尚玲, 韩渊怀, 霍轶琼, 侯思宇. 基于代谢组学研究谷子SiYABBYs参与花发育过程中鼠李糖苷的生物合成[J]. 生物技术通报, 2023, 39(6): 189-198. |
| [3] | 韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. |
| [4] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [5] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [6] | 位欣欣, 兰海燕. 植物MYB转录因子调控次生代谢及逆境响应的研究进展[J]. 生物技术通报, 2022, 38(8): 12-23. |
| [7] | 张婵, 吴友根, 于靖, 杨东梅, 姚广龙, 杨华庚, 张军锋, 陈萍. 光与茉莉酸信号介导的萜类化合物合成分子机制[J]. 生物技术通报, 2022, 38(8): 32-40. |
| [8] | 李萍, 郭发平, 田敏, 税阳, 徐娜娜, 白大嵩, 余德金, 张杰, 胡运高, 彭友林. 甾醇在调节植物生长发育中的研究进展[J]. 生物技术通报, 2022, 38(7): 90-98. |
| [9] | 古丽加马力·艾萨, 邢军, 李安, 张瑞. 开菲尔粒中微生物对苯并(α)芘的非靶向代谢组学分析[J]. 生物技术通报, 2022, 38(5): 123-135. |
| [10] | 林科运, 段钰晶, 王高升, 孙念礼, 方玉洁, 王幼平. 甘蓝型油菜BnNF-YA1的克隆和功能鉴定[J]. 生物技术通报, 2022, 38(4): 106-116. |
| [11] | 孙曼銮, 葛赛, 卜佳, 朱壮彦. 大肠杆菌核糖核酸酶调控机制研究[J]. 生物技术通报, 2022, 38(3): 234-245. |
| [12] | 杨玉萍, 张霞, 王翀翀, 王晓艳. 不同年龄大鼠尿液代谢组学研究[J]. 生物技术通报, 2022, 38(2): 166-172. |
| [13] | 悦曼芳, 张春, 吴忠义. 植物转录因子AP2/ERF家族蛋白结构和功能的研究进展[J]. 生物技术通报, 2022, 38(12): 11-26. |
| [14] | 汤晓丽, 姜福东, 张洪霞. 植物SINA E3泛素连接酶功能的研究进展[J]. 生物技术通报, 2022, 38(10): 10-17. |
| [15] | 山琦, 贾惠舒, 姚文博, 刘伟灿, 李海燕. 植物miR396-GRF模块的生物学功能及其潜在应用价值[J]. 生物技术通报, 2022, 38(10): 34-44. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
