








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 189-198.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1463
韩华蕊1( ), 杨宇琭1, 门艺涵1, 韩尚玲1, 韩渊怀1,2,3, 霍轶琼1,2(
), 杨宇琭1, 门艺涵1, 韩尚玲1, 韩渊怀1,2,3, 霍轶琼1,2( ), 侯思宇1,2,3(
), 侯思宇1,2,3( )
)
收稿日期:2022-11-29
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
侯思宇,男,副教授,研究方向:种质创新与遗传工程;E-mail: bragren123@163.com;作者简介:韩华蕊,女,硕士研究生,研究方向:种质创新与遗传工程;E-mail: 2594488988@qq.com
基金资助:
HAN Hua-rui1( ), YANG Yu-lu1, MEN Yi-han1, HAN Shang-ling1, HAN Yuan-huai1,2,3, HUO Yi-qiong1,2(
), YANG Yu-lu1, MEN Yi-han1, HAN Shang-ling1, HAN Yuan-huai1,2,3, HUO Yi-qiong1,2( ), HOU Si-yu1,2,3(
), HOU Si-yu1,2,3( )
)
Received:2022-11-29
Published:2023-06-26
Online:2023-07-07
摘要:
花器官发育是生殖发育的前提和基础,对小穗最终的形成具有决定性作用,YABBY转录因子在花器官的发育中具有重要作用,探究谷子YABBY家族成员的序列特征及表达模式,揭示其与谷子穗发育的关系,为阐明谷子YABBY基因家族的功能以及提高谷子产量奠定理论基础。通过对谷子YABBY基因家族进行全基因组鉴定,分析SiYABBYs的理化性质、组织表达模式,并与拟南芥和水稻YABBY蛋白序列比对进行功能预测。结果表明,共鉴定8个YABBY家族成员,编码150-303个氨基酸,在抽穗期和授粉期的穗中特异表达,可能参与谷子营养组织的发育和花器官的形成。对谷子抽穗期和授粉期的代谢组数据与SiYABBYs的表达水平进行相关性分析发现,SiYABBY1/SiYABBY5/SiYABBY7/SiYABBY8与鼠李糖苷代谢通路的酶基因紧密关联,表明促进授粉期穗中鼠李糖苷的积累,可能与谷子的成育率及结实率相关。
韩华蕊, 杨宇琭, 门艺涵, 韩尚玲, 韩渊怀, 霍轶琼, 侯思宇. 基于代谢组学研究谷子SiYABBYs参与花发育过程中鼠李糖苷的生物合成[J]. 生物技术通报, 2023, 39(6): 189-198.
HAN Hua-rui, YANG Yu-lu, MEN Yi-han, HAN Shang-ling, HAN Yuan-huai, HUO Yi-qiong, HOU Si-yu. SiYABBYs Involved in Rhamnoside Biosynthesis During the Flower Development of Setaria italica, Based on Metabolomics[J]. Biotechnology Bulletin, 2023, 39(6): 189-198.
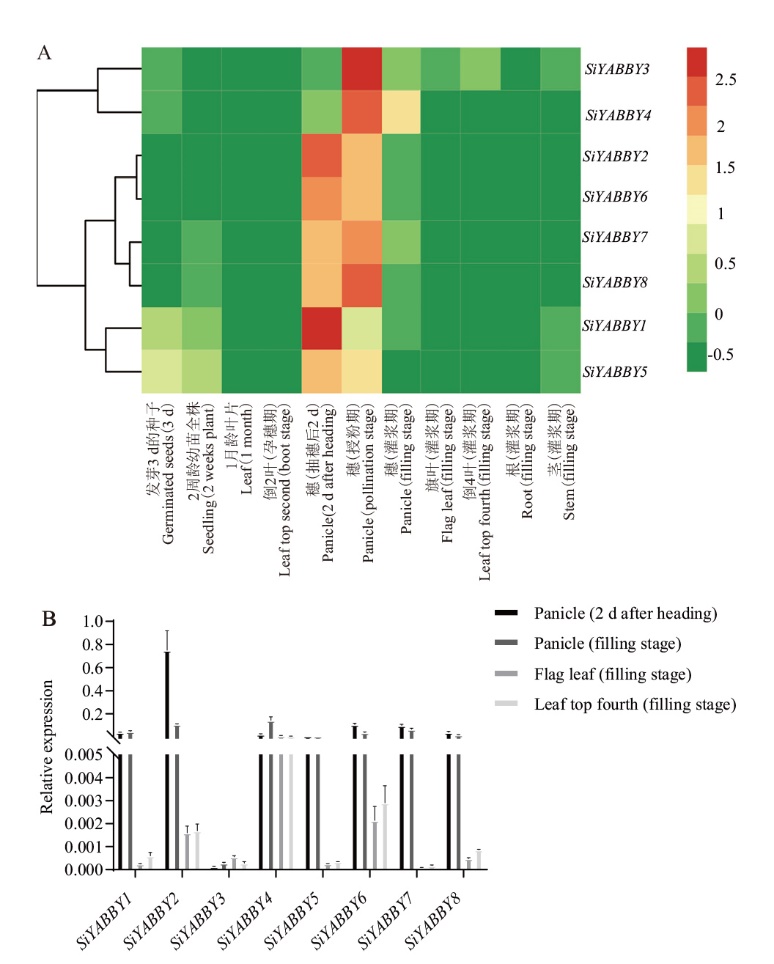
图1 SiYABBYs家族成员组织表达谱 A:谷子SiYABBY在xiaomi不同组织不同时期的组织表达谱;B:RT-qPCR结果
Fig. 1 Expression profile of SiYABBYs A: Tissue expression profile of SiYABBY gene in different tissues and different periods of xiaomi. B: The result of RT-qPCR
| 亚族 Subfamily | 在拟南芥中的功能 Function in Arabidopsis thaliana | 在水稻中的功能 Function in rice | 谷子中功能预测 Function prediction in foxtail millet |
|---|---|---|---|
| YAB3 | AtFIL:调控花序和花分生组织的形成和发育 ATFIL: Regulates the formation and development of inflorescence and floral meristems | OsYABBY3:侧生器官的形成和分生组织的分化 OsYABBY3: Formation of lateral organs and differentiation of meristems | SiYABBY1/SiYABBY5/SiYABBY7:参与侧生器官的发育以及花器官和小穗的形成和发育 SiYABBY1/SiYABBY5/SiYABBY7: Participate in lateral organ development and the formation and development of flower organs and spikelet |
| AtYAB3:参与营养组织的发育 AtYAB3: Participate in the development of nutritive tissues | OsYABBY4:调控维管组织的发育和叶片的极性 OsYABBY4: Regulates vascular tissue development and leaf polarity | ||
| OsYABBY5:小穗发育 OsYABBY5: Spikelets development | |||
| INO | AtINO:调节外胚珠珠被的发育 AtINO: To regulate the development of the outer ovules | OsYABBY7 | SiYABBY3:调节外胚珠和花器官的发育 SiYABBY3: Adjust the ovule and floral organ development |
| CRC | AtCRC:心皮发育的关键调节因子 AtCRC: Carpel development a key regulating factor | OsDL:调控心皮的形成 OsDL: Regulate the formation of carpels | SiYABBY8(SiDL):导致开花延迟、叶片卷曲和种子变小以及参与心皮的形成 SiYABBY8(SiDL): Cause delayed flowering, smaller hairy leaves and seeds, and participate in the formation of carpel |
| YAB2 | AtYAB2:参与营养组织的发育 AtYAB2: Participate in the development of nutritive tissues | OsYABBY1:雄蕊和心皮的分生组织发育 OsYABBY1: Meristematic development of stamens and carpels | SiYABBY2/SiYABBY4/SiYABBY6:参与营养组织的发育和花器官的形成 SiYABBY2/SiYABBY4/SiYABBY6: Involved in the development of vegetative tissues and the formation of floral organs |
| OsYABBY2 | |||
| OsYABBY6:调控水稻的株高和产量 OsYABBY6: Regulate the plant height and yield of rice |
表1 谷子SiYABBYs的功能预测
Table 1 Functional prediction of SiYABBYs in foxtail millet
| 亚族 Subfamily | 在拟南芥中的功能 Function in Arabidopsis thaliana | 在水稻中的功能 Function in rice | 谷子中功能预测 Function prediction in foxtail millet |
|---|---|---|---|
| YAB3 | AtFIL:调控花序和花分生组织的形成和发育 ATFIL: Regulates the formation and development of inflorescence and floral meristems | OsYABBY3:侧生器官的形成和分生组织的分化 OsYABBY3: Formation of lateral organs and differentiation of meristems | SiYABBY1/SiYABBY5/SiYABBY7:参与侧生器官的发育以及花器官和小穗的形成和发育 SiYABBY1/SiYABBY5/SiYABBY7: Participate in lateral organ development and the formation and development of flower organs and spikelet |
| AtYAB3:参与营养组织的发育 AtYAB3: Participate in the development of nutritive tissues | OsYABBY4:调控维管组织的发育和叶片的极性 OsYABBY4: Regulates vascular tissue development and leaf polarity | ||
| OsYABBY5:小穗发育 OsYABBY5: Spikelets development | |||
| INO | AtINO:调节外胚珠珠被的发育 AtINO: To regulate the development of the outer ovules | OsYABBY7 | SiYABBY3:调节外胚珠和花器官的发育 SiYABBY3: Adjust the ovule and floral organ development |
| CRC | AtCRC:心皮发育的关键调节因子 AtCRC: Carpel development a key regulating factor | OsDL:调控心皮的形成 OsDL: Regulate the formation of carpels | SiYABBY8(SiDL):导致开花延迟、叶片卷曲和种子变小以及参与心皮的形成 SiYABBY8(SiDL): Cause delayed flowering, smaller hairy leaves and seeds, and participate in the formation of carpel |
| YAB2 | AtYAB2:参与营养组织的发育 AtYAB2: Participate in the development of nutritive tissues | OsYABBY1:雄蕊和心皮的分生组织发育 OsYABBY1: Meristematic development of stamens and carpels | SiYABBY2/SiYABBY4/SiYABBY6:参与营养组织的发育和花器官的形成 SiYABBY2/SiYABBY4/SiYABBY6: Involved in the development of vegetative tissues and the formation of floral organs |
| OsYABBY2 | |||
| OsYABBY6:调控水稻的株高和产量 OsYABBY6: Regulate the plant height and yield of rice |
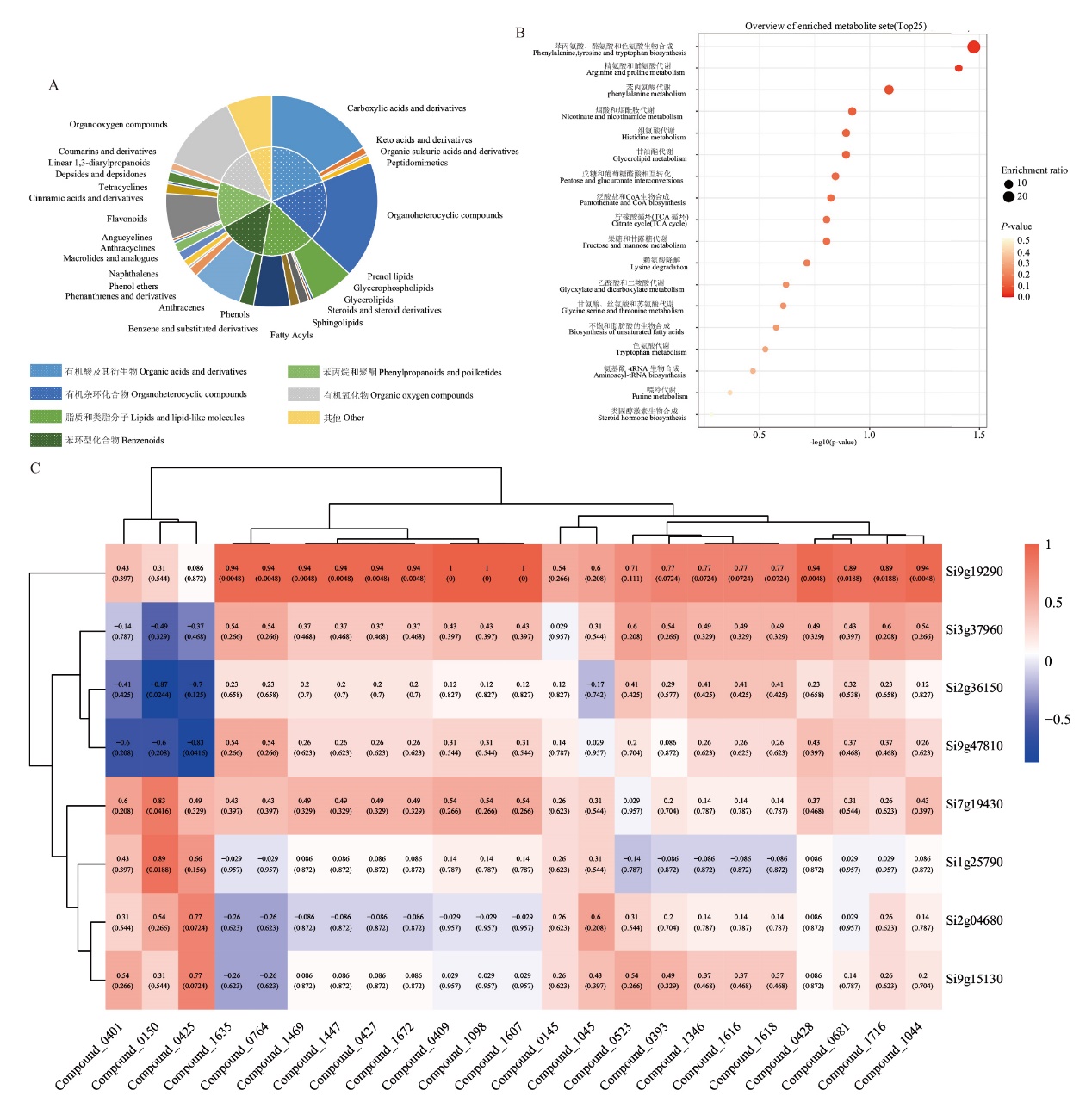
图2 xiaomi不同样品中差异代谢物分析 A:差异代谢物分类;B:差异代谢物富集分析;C:苯丙烷和聚酮类化合物与SiYABBY的相关性分析
Fig. 2 Differential metabolites analysis in different xiaomi samples A: Differences metabolites classification. B: Differential metabolite enrichment analysis. C: Correlation analysis of phenylpropane and polyketones in differential metabolites with SiYABBY
| 转录因子 Transcription factor | 结合基序 Motif ID | 靶基因 Target gene | 模板链 Strand | 起始位置 Start | 终止为止 End | P值 P-value | q值 q-value | 匹配序列 Matched sequence |
|---|---|---|---|---|---|---|---|---|
| SiYABBY8 | UN0396.1 | Si1g24720 | - | 1 734 | 1 741 | 2.65e-05 | 1 | TAATCATA |
| Si1g07250 | + | 698 | 705 | 5.3e-05 | 1 | TTATCATA | ||
| Si1g24750 | + | 1 165 | 1 172 | 7.95e-05 | 1 | TAATCATA | ||
| Si7g05130 | + | 1 478 | 1 485 | 7.95e-05 | 1 | TAATCATT | ||
| SiYABBY1 SiYABBY5 SiYABBY7 | UN0415.1 | Si8g14530 | - | 121 | 128 | 3.23e-05 | 0.529 | AATAATAA |
| Si9g17990 | - | 1 088 | 1 095 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g30480 | + | 1 319 | 1 326 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si9g30300 | - | 1 314 | 1 321 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g01180 | - | 1 785 | 1 792 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g01180 | - | 389 | 396 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si6g16950 | - | 861 | 868 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si6g16950 | + | 896 | 903 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g30480 | - | 1 593 | 1 600 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si9g04210 | + | 1 645 | 1 652 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si9g04210 | - | 1 647 | 1 654 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g01180 | - | 1 793 | 1 800 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g05130 | - | 164 | 171 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si9g52070 | + | 249 | 256 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si7g01180 | - | 431 | 438 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si9g30300 | + | 849 | 856 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si7g30480 | - | 1 452 | 1 459 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si5g36410 | + | 1 437 | 1 444 | 9.69e-05 | 0.466 | AATATTAA |
表2 鼠李糖苷代谢通路酶基因与SiYABBY转录因子结合基序分析
Table 2 The analysis of binding motif between rhamnoside metabolic pathway enzyme genes and SiYABBY transcription factor
| 转录因子 Transcription factor | 结合基序 Motif ID | 靶基因 Target gene | 模板链 Strand | 起始位置 Start | 终止为止 End | P值 P-value | q值 q-value | 匹配序列 Matched sequence |
|---|---|---|---|---|---|---|---|---|
| SiYABBY8 | UN0396.1 | Si1g24720 | - | 1 734 | 1 741 | 2.65e-05 | 1 | TAATCATA |
| Si1g07250 | + | 698 | 705 | 5.3e-05 | 1 | TTATCATA | ||
| Si1g24750 | + | 1 165 | 1 172 | 7.95e-05 | 1 | TAATCATA | ||
| Si7g05130 | + | 1 478 | 1 485 | 7.95e-05 | 1 | TAATCATT | ||
| SiYABBY1 SiYABBY5 SiYABBY7 | UN0415.1 | Si8g14530 | - | 121 | 128 | 3.23e-05 | 0.529 | AATAATAA |
| Si9g17990 | - | 1 088 | 1 095 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g30480 | + | 1 319 | 1 326 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si9g30300 | - | 1 314 | 1 321 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g01180 | - | 1 785 | 1 792 | 3.23e-05 | 0.529 | AATAATAA | ||
| Si7g01180 | - | 389 | 396 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si6g16950 | - | 861 | 868 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si6g16950 | + | 896 | 903 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g30480 | - | 1 593 | 1 600 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si9g04210 | + | 1 645 | 1 652 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si9g04210 | - | 1 647 | 1 654 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g01180 | - | 1 793 | 1 800 | 6.46e-05 | 0.529 | AATAATTA | ||
| Si7g05130 | - | 164 | 171 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si9g52070 | + | 249 | 256 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si7g01180 | - | 431 | 438 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si9g30300 | + | 849 | 856 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si7g30480 | - | 1 452 | 1 459 | 9.69e-05 | 0.56 | AATATTAA | ||
| Si5g36410 | + | 1 437 | 1 444 | 9.69e-05 | 0.466 | AATATTAA |
| [1] |
韩尚玲, 霍轶琼, 等. 基于WGCNA发掘谷子穗部类黄酮合成途径调控关键基因[J]. 作物学报, 2022, 48(7): 1645-1657.
doi: 10.3724/SP.J.1006.2022.14107 |
| Han SL, Huo YQ, et al. Identification of regulatory genes related to flavonoids synthesis by weighted gene correlation network analysis in the panicle of foxtail millet[J]. Acta Agron Sin, 2022, 48(7): 1645-1657. | |
| [2] |
Yang ZR, Zhang HS, Li XK, et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system[J]. Nat Plants, 2020, 6(9): 1167-1178.
doi: 10.1038/s41477-020-0747-7 |
| [3] | 顾伟航. 水稻花器官发育分子调控机制的研究[D]. 上海: 上海交通大学, 2018. |
| Gu WH. The molecular mechanisms of rice flower development[D]. Shanghai: Shanghai Jiao Tong University, 2018. | |
| [4] |
Zhu Y, Wagner D. Plant inflorescence architecture: the formation, activity, and fate of axillary meristems[J]. Cold Spring Harb Perspect Biol, 2020, 12(1): a034652.
doi: 10.1101/cshperspect.a034652 URL |
| [5] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [6] |
Romanova MA, Maksimova AI, Pawlowski K, et al. YABBY genes in the development and evolution of land plants[J]. Int J Mol Sci, 2021, 22(8): 4139.
doi: 10.3390/ijms22084139 URL |
| [7] |
Golz JF, Roccaro M, Kuzoff R, et al. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves[J]. Development, 2004, 131(15): 3661-3670.
doi: 10.1242/dev.01221 URL |
| [8] |
Zhang TP, Li CY, Li DX, et al. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants[J]. J Plant Res, 2020, 133(6): 751-763.
doi: 10.1007/s10265-020-01227-7 pmid: 33033876 |
| [9] |
Sawa S, Ito T, Shimura Y, et al. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems[J]. Plant Cell, 1999, 11(1): 69-86.
pmid: 9878633 |
| [10] |
Lee JY, Baum SF, Oh SH, et al. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade[J]. Development, 2005, 132(22): 5021-5032.
doi: 10.1242/dev.02067 URL |
| [11] | Shchennikova AV, Slugina MA, Beletsky AV, et al. The YABBY genes of leaf and leaf-like organ polarity in leafless plant Monotropa hypopitys[J]. Int J Genomics, 2018, 2018: 7203469. |
| [12] |
Villanueva JM, Broadhvest J, Hauser BA, et al. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules[J]. Genes Dev, 1999, 13(23): 3160-3169.
doi: 10.1101/gad.13.23.3160 URL |
| [13] |
Yamaguchi T, Nagasawa N, et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa[J]. Plant Cell, 2004, 16(2): 500-509.
pmid: 14729915 |
| [14] |
Tanaka W, Toriba T, Ohmori Y, et al. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet[J]. Plant Cell, 2012, 24(1): 80-95.
doi: 10.1105/tpc.111.094797 URL |
| [15] |
Jang S, Hur J, Kim SJ, et al. Ectopic expression of OsYAB1causes extra stamens and carpels in rice[J]. Plant Mol Biol, 2004, 56(1): 133-143.
doi: 10.1007/s11103-004-2648-y URL |
| [16] |
Dai MQ, Hu YF, Zhao Y, et al. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development[J]. Plant Physiol, 2007, 144(1): 380-390.
doi: 10.1104/pp.107.095737 URL |
| [17] | 夏妙林, 唐冬英, 杨远柱, 等. 水稻OsYABBY6基因参与叶片发育的初步研究[J]. 生命科学研究, 2017, 21(1): 23-30. |
| Xia ML, Tang DY, Yang YZ, et al. Preliminary study on the rice OsYABBY6 gene involving in the regulation of leaf development[J]. Life Sci Res, 2017, 21(1): 23-30. | |
| [18] |
Strable J, Wallace JG, Unger-Wallace E, et al. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture[J]. Plant Cell, 2017, 29(7): 1622-1641.
doi: 10.1105/tpc.16.00477 URL |
| [19] |
Juarez MT, Twigg RW, Timmermans MCP. Specification of adaxial cell fate during maize leaf development[J]. Development, 2004, 131(18): 4533-4544.
doi: 10.1242/dev.01328 pmid: 15342478 |
| [20] |
赵翔宇, 谢洪涛, 陈祥彬, 等. 小麦TaYAB2基因的过量表达造成转基因拟南芥叶片近轴面特征趋向远轴面[J]. 作物学报, 2012, 38(11): 2042-2051.
doi: 10.3724/SP.J.1006.2012.02042 |
|
Zhao XY, Xie HT, Chen XB, et al. Ectopic expression of TaYAB2, a member of YABBY gene family in wheat, causes partial abaxialization of adaxial epidermises of leaves in Arabidopsis[J]. Acta Agron Sin, 2012, 38(11): 2042-2051.
doi: 10.3724/SP.J.1006.2012.02042 URL |
|
| [21] |
Guo J, Zhou X, Dai K, et al. Comprehensive analysis of YABBY gene family in foxtail millet(Setaria italica)and functional characterization of SiDL[J]. J Integr Agric, 2022, 21(10): 2876-2887.
doi: 10.1016/j.jia.2022.07.052 URL |
| [22] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [23] |
Yamada T, Yokota S, Hirayama Y, et al. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms[J]. Plant J, 2011, 67(1): 26-36.
doi: 10.1111/tpj.2011.67.issue-1 URL |
| [24] |
Bowman JL. The YABBY gene family and abaxial cell fate[J]. Curr Opin Plant Biol, 2000, 3(1): 17-22.
doi: 10.1016/s1369-5266(99)00035-7 pmid: 10679447 |
| [25] |
Gross T, Broholm S, Becker A. CRABS CLAW acts as a bifunctional transcription factor in flower development[J]. Front Plant Sci, 2018, 9: 835.
doi: 10.3389/fpls.2018.00835 pmid: 29973943 |
| [26] |
Ikeda T, Tanaka W, Mikami M, et al. Generation of artificial drooping leaf mutants by CRISPR-Cas9 technology in rice[J]. Genes Genet Syst, 2016, 90(4): 231-235.
doi: 10.1266/ggs.15-00030 pmid: 26617267 |
| [27] |
Ishikawa M, Ohmori Y, Tanaka W, et al. The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses[J]. Genes Genet Syst, 2009, 84(2): 137-146.
doi: 10.1266/ggs.84.137 URL |
| [28] |
Stahle MI, Kuehlich J, Staron L, et al. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis[J]. Plant Cell, 2009, 21(10): 3105-3118.
doi: 10.1105/tpc.109.070458 URL |
| [29] |
Dai MQ, Hu YF, Zhao Y, et al. Regulatory networks involving YABBY genes in rice shoot development[J]. Plant Signal Behav, 2007, 2(5): 399-400.
doi: 10.4161/psb.2.5.4279 URL |
| [30] |
Yang C, Ma YM, Li JX. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway[J]. J Exp Bot, 2016, 67(18): 5545-5556.
doi: 10.1093/jxb/erw319 URL |
| [31] | Dai MQ, Zhao Y, Ma Q, et al. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism[J]. Plant Physiol, 2007, 144(1): 121-133. |
| [32] |
Yamada T, et al. Expression pattern of INNER NO OUTER homologue in Nymphaea(Water Lily Family, Nymphaeaceae)[J]. Dev Genes Evol, 2003, 213(10): 510-513.
doi: 10.1007/s00427-003-0350-8 URL |
| [33] | Karioti A, Kitsaki CK, Zygouraki S, et al. Occurrence of flavonoids in Ophrys(Orchidaceae)flower parts[J]. Flora Morphol Distribution Funct Ecol Plants, 2008, 203(7): 602-609. |
| [34] |
Zhou CH, Sun CD, Chen KS, et al. Flavonoids, phenolics, and antioxidant capacity in the flower of Eriobotrya japonica Lindl[J]. Int J Mol Sci, 2011, 12(5): 2935-2945.
doi: 10.3390/ijms12052935 URL |
| [35] |
Shen N, Wang T, Gan Q, et al. Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity[J]. Food Chem, 2022, 383: 132531.
doi: 10.1016/j.foodchem.2022.132531 URL |
| [36] | Amalesh S, Gouranga D, Sanjoy KD. Roles of flavonoids in plants[J]. Int J Pharm Sci Tech, 2011, 6(1): 12-35. |
| [1] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| [2] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [3] | 邢媛, 宋健, 李俊怡, 郑婷婷, 刘思辰, 乔治军. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251. |
| [4] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [5] | 古丽加马力·艾萨, 邢军, 李安, 张瑞. 开菲尔粒中微生物对苯并(α)芘的非靶向代谢组学分析[J]. 生物技术通报, 2022, 38(5): 123-135. |
| [6] | 杨玉萍, 张霞, 王翀翀, 王晓艳. 不同年龄大鼠尿液代谢组学研究[J]. 生物技术通报, 2022, 38(2): 166-172. |
| [7] | 吴玉苹, 周勇, 蒲娟, 李会, 章金刚, 朱艳平. 代谢组学在肿瘤药物靶点筛选中的应用进展[J]. 生物技术通报, 2022, 38(1): 311-318. |
| [8] | 张凤, 陈伟. 代谢组学在植物逆境生物学中的研究进展[J]. 生物技术通报, 2021, 37(8): 1-11. |
| [9] | 陈倩, 张露源, 陈伯昌, 吴海燕. 大豆孢囊线虫生防菌株Myrothecium verrucaria ZW-2发酵条件优化及活性物质分析[J]. 生物技术通报, 2021, 37(7): 127-136. |
| [10] | 田鹤, 税光厚. 基于质谱技术的代谢组学分析方法研究进展[J]. 生物技术通报, 2021, 37(1): 24-32. |
| [11] | 殷志斌, 黄文洁, 伍欣宙, 晏石娟. 空间分辨代谢组学进展和挑战[J]. 生物技术通报, 2021, 37(1): 32-51. |
| [12] | 黄小丹, 陈梦雨, 黄文洁, 张名位, 晏石娟. 基于代谢组学的植物多酚及其肠道健康效应研究进展[J]. 生物技术通报, 2021, 37(1): 123-136. |
| [13] | 李锐, 孙祖莉, 杨贤庆, 李来好, 魏涯, 岑剑伟, 王晶, 赵永强. 代谢组学在水产品品质与安全中的研究进展[J]. 生物技术通报, 2020, 36(11): 155-163. |
| [14] | 兰青阔, 赵新, 沈晓玲, 魏静娜, 刘双, 陈锐, 檀建新, 王永. 基于代谢组学的转基因水稻生物安全评价方法研究[J]. 生物技术通报, 2020, 36(11): 222-229. |
| [15] | 赖博文, 刘玢, 梁永康. 基于高分辨质谱的非靶向代谢组学在食品造假鉴定中的研究进展[J]. 生物技术通报, 2019, 35(2): 192-197. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||