








生物技术通报 ›› 2022, Vol. 38 ›› Issue (5): 149-158.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1067
赵明明1( ), 唐殷1, 郭磊周1, 韩佳慧1, 葛佳茗1, 孟勇2, 平淑珍1, 周正富1, 王劲1(
), 唐殷1, 郭磊周1, 韩佳慧1, 葛佳茗1, 孟勇2, 平淑珍1, 周正富1, 王劲1( )
)
收稿日期:2021-08-09
出版日期:2022-05-26
发布日期:2022-06-10
作者简介:赵明明,女,硕士研究生,研究方向:微生物分子生物学与基因工程;E-mail: 基金资助:
ZHAO Ming-ming1( ), TANG Yin1, GUO Lei-zhou1, HAN Jia-hui1, GE Jia-ming1, MENG Yong2, PING Shu-zhen1, ZHOU Zheng-fu1, WANG Jin1(
), TANG Yin1, GUO Lei-zhou1, HAN Jia-hui1, GE Jia-ming1, MENG Yong2, PING Shu-zhen1, ZHOU Zheng-fu1, WANG Jin1( )
)
Received:2021-08-09
Published:2022-05-26
Online:2022-06-10
摘要:
Lon是一类广泛存在于生物体内的蛋白水解酶,其通过水解受损及无用蛋白,防止蛋白聚集造成的细胞伤害,在胞内氨基酸周转再利用中发挥重要作用。极端微生物耐辐射异常球菌(Deinococcus radiodurans R1)的超强非生物胁迫耐受性与其拥有功能强大的胞内蛋白质稳态系统密切相关。而Lon作为蛋白水解系统的重要组成部分,在耐辐射异常球菌中的功能分析还未见具体报导。因此本研究以耐辐射异常球菌Lon1蛋白酶为研究对象,分析其逆境胁迫下的功能。结果显示 lon1基因的转录受高温诱导,48℃高温胁迫蛋白组学分析显示,lon1的缺失使多个分子伴侣蛋白、参与膜功能和转运蛋白以及与DNA修复、转录调控、能量代谢等相关的蛋白表达量发生显著上调;lon1的缺失使5个参与细胞形态建成的蛋白表达量显著下调,电镜结果证实lon1缺失使细菌分裂异常,形成巨型细胞,细胞膜发生严重的损伤;非生物胁迫冲击实验发现,lon1的缺失使耐辐射异常球菌对0.2 mol/L NaCl以及48℃高温敏感。研究表明Lon1蛋白酶主要参与耐辐射异常球菌的细胞形态建成,并在适应高温等胁迫中发挥功能。
赵明明, 唐殷, 郭磊周, 韩佳慧, 葛佳茗, 孟勇, 平淑珍, 周正富, 王劲. Lon1蛋白酶参与耐辐射异常球菌高温胁迫及细胞分裂的功能研究[J]. 生物技术通报, 2022, 38(5): 149-158.
ZHAO Ming-ming, TANG Yin, GUO Lei-zhou, HAN Jia-hui, GE Jia-ming, MENG Yong, PING Shu-zhen, ZHOU Zheng-fu, WANG Jin. Function Analysis of Lon1 Protease Involved in High Temperature Stress and Cell Division of Deinococcus radiodurans R1[J]. Biotechnology Bulletin, 2022, 38(5): 149-158.
| 菌株及质粒Strains and plasmids | 特性Relevant characteristic | 来源Source | |
|---|---|---|---|
| 菌株 | Deinococcus radioduransR1 | 耐辐射异常球菌野生型 | 中科院微生物所菌种保藏中心 |
| △lon1 | 缺失lon1基因的突变株(Kanr) | 本研究构建 | |
| comlon1 | lon1回补株(Kanr+ Cmr) | 本研究构建 | |
| Z3-△lon1 | △lon1中导入质粒pRADZ3(Cmr) | 本研究构建 | |
| 质粒 | pRAD Z3 | 构建回补菌株所用质粒 | 本实验室保存 |
表1 菌株与质粒
Table 1 Strains and Plasmids
| 菌株及质粒Strains and plasmids | 特性Relevant characteristic | 来源Source | |
|---|---|---|---|
| 菌株 | Deinococcus radioduransR1 | 耐辐射异常球菌野生型 | 中科院微生物所菌种保藏中心 |
| △lon1 | 缺失lon1基因的突变株(Kanr) | 本研究构建 | |
| comlon1 | lon1回补株(Kanr+ Cmr) | 本研究构建 | |
| Z3-△lon1 | △lon1中导入质粒pRADZ3(Cmr) | 本研究构建 | |
| 质粒 | pRAD Z3 | 构建回补菌株所用质粒 | 本实验室保存 |
| 扩增产物 Product for PCR amplification | 引物名称 Primer name | 序列Sequence(5'-3') | 片段长度Size/bp |
|---|---|---|---|
| lon1-Up | lon1-UP-F | AGCTGCTGACCCGTGACACCGAGCCATGGAGACCGAGGGCCC | 613 |
| lon1-up-R | GGGCCCTCGGTCTCCATGGCTCGGTGTCACGGGTCAGCAGCT | ||
| lon1-Kan | Kan-lon1-F | AGCTGCTGACCCGTGACACCGAGCCATGGAGACCGAGGGCCC | 965 |
| Kan-Down-R | TGGTCCACGCCAGGCCCTGCGTTAGAAAAACTCATCGAGCAT | ||
| lon1-Down | lon1-Down-F | ATGCTCGATGAGTTTTTCTAACGCAGGGCCTGGCGTGGACCA | 614 |
| lon1-Down-R | TTATGCGCCCGGCTGCACCGA | ||
| Kan内部序列 | YZ-Km-F | CGATTGTATGGGAAGCCCGAT | 591 |
| YZ-Km-R | CTCACCGAGGCAGTTCCATAG | ||
| 敲除的lon1序列 | YZ-lon1-F | ACCGTGGTTCGCAACTACAT | 400 |
| YZ-lon1-R | CGATCTCGACCTTCTCCTGC |
表2 引物序列
Table 2 Sequences of Primers
| 扩增产物 Product for PCR amplification | 引物名称 Primer name | 序列Sequence(5'-3') | 片段长度Size/bp |
|---|---|---|---|
| lon1-Up | lon1-UP-F | AGCTGCTGACCCGTGACACCGAGCCATGGAGACCGAGGGCCC | 613 |
| lon1-up-R | GGGCCCTCGGTCTCCATGGCTCGGTGTCACGGGTCAGCAGCT | ||
| lon1-Kan | Kan-lon1-F | AGCTGCTGACCCGTGACACCGAGCCATGGAGACCGAGGGCCC | 965 |
| Kan-Down-R | TGGTCCACGCCAGGCCCTGCGTTAGAAAAACTCATCGAGCAT | ||
| lon1-Down | lon1-Down-F | ATGCTCGATGAGTTTTTCTAACGCAGGGCCTGGCGTGGACCA | 614 |
| lon1-Down-R | TTATGCGCCCGGCTGCACCGA | ||
| Kan内部序列 | YZ-Km-F | CGATTGTATGGGAAGCCCGAT | 591 |
| YZ-Km-R | CTCACCGAGGCAGTTCCATAG | ||
| 敲除的lon1序列 | YZ-lon1-F | ACCGTGGTTCGCAACTACAT | 400 |
| YZ-lon1-R | CGATCTCGACCTTCTCCTGC |
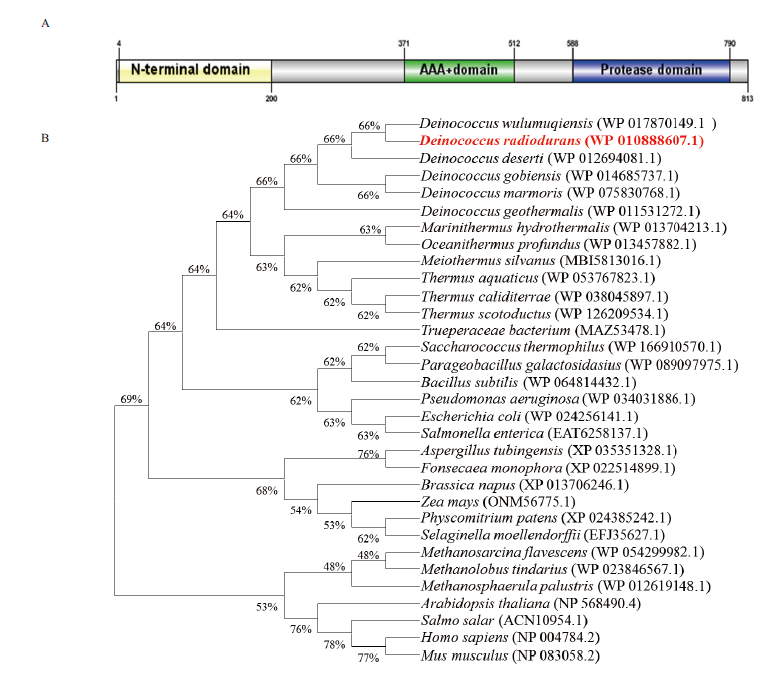
图1 Lon1结构域及系统发育分析 A:48℃高温处理;B:UV处理;C:80 mmol/L H2O2处理;D:0.2 mol/L NaCl处理
Fig. 1 Structural domain and phylogenetic analysis of Lon1 A:48℃ heat;B:UV;C:80 mmol/L H2O2;D:0.2 mol/L NaCl
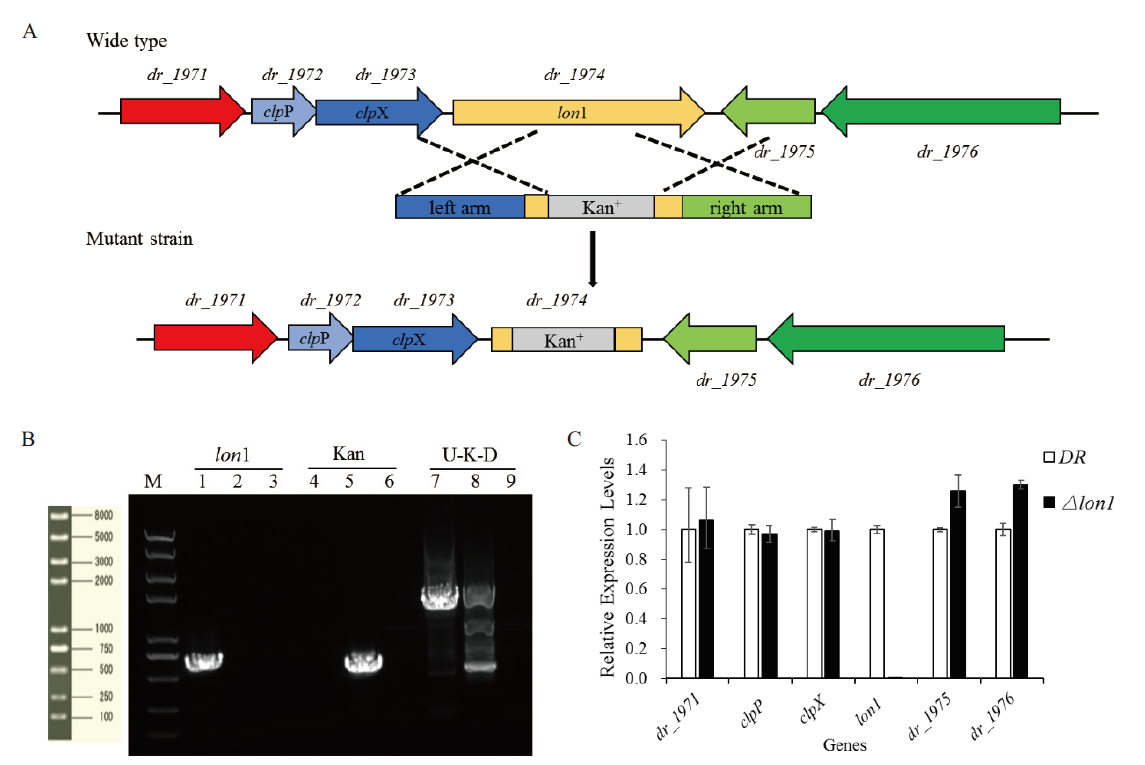
图3 lon1突变株的构建与验证 A:构建lon1突变株示意图;B:验证lon1突变株电泳图(M:Trans 2K plus Ⅱ;1,4,7 野生型WT的PCR产物;2,5,8:突变株lon1的PCR产物;3,6,9:阴性对照);C:lon1缺失后对临近基因转录水平的影响
Fig. 3 Construction and identification of the lon1 mutant strain A:Schematic representation of the Δlon1mutant. B:Electrophoretogram of the lon1 mutant strain(M:Trans 2K PlusⅡ DNA marker. 1, 4, 7:PCR products amplified from wild type D. radiodurans. 2, 5, 8:PCR products amplified from the lon1mutant. 3, 6, 9:Negative control). C:Effects of lon1 deleted on adjacent genes in transcriptional level
| UniProtKB | Locus | Predicted function | Protein name | FC |
|---|---|---|---|---|
| Protein fate(heat shock responsive chaperones and protease) | ||||
| Q9RV58 | DR_1172 | Chaperone-like protein | DosH | 13.37 |
| Q9RWQ9 | DR_0607 | 60 kD chaperone | GroL | 7.08 |
| Q9RVI3 | DR_1046 | Chaperone protein ClpB | ClpB | 4.70 |
| Q9RXG4 | DR_0349 | Lon protease | Lon2 | 4.52 |
| Q9RWR0 | DR_0606 | 10 kD chaperone | GroS | 4.46 |
| Q9RY24 | DR_0128 | Chaperone protein GrpE | GrpE | 3.24 |
| DNA metabolism(repair and recombination) | ||||
| Q9RXI7 | DR_0326 | DNA damage response protein D | DdrD | 2.59 |
| Q9RS64 | DR_2263 | DNA protection during starvation protein 1 | Dps1 | 2.88 |
| Q9RY51 | DR_0099 | Single-stranded DNA-binding protein | Ssb | 3.25 |
| Q9RSJ6 | DR_2128 | DNA-directed RNA polymerase subunit alpha | RpoA | 2.06 |
| Q9RRE4 | DR_2548 | Transcriptional regulatory protein | 7.75 | |
| Membrane functions and transport | ||||
| Q9RWU0 | DR_0575 | Protein translocase subunit SecA | SecA | 2.18 |
| P56867 | DR_2508 | Hexagonally packed intermediate-layer surface protein | Hpi | 3.48 |
| Q9RWH3 | DR_0695 | V-type ATP synthase subunit | AtpI | 1.57 |
| Translation | ||||
| Q9RST0 | DR_2043 | 50S ribosomal protein L7/L12 | RplL | 0.14 |
| Central intermediary metabolism(synthesis of nitrogen,sulfur,polyamine compounds) | ||||
| Q9RR70 | DR_2627 | Fumarate hydratase class II | FumC | 7.59 |
| Q9RWB2 | DR_0757 | Citrate synthase | GltA | 3.06 |
| Q9RZ06 | DR_A0147 | Histidine ammonia-lyase | HutH | 2.66 |
| Q9RTN7 | DR_1720 | Aconitate hydratase A | Acn | 2.10 |
| P56861 | DR_A0014 | Adenylyl-sulfate kinase | CysC | 0.23 |
| Amino acid biosynthesis and energy metabolism | ||||
| Q9RRT0 | DR_2405 | Demethylmenaquinone methyltransferase | MenG | 0.51 |
| Q9RV69 | DR_1161 | 5-hydroxyisourate hydrolase | 0.33 | |
| Q9RV98 | DR_1131 | Ferrochelatase | HemH | 2.04 |
| Q9RRC4 | DR_2568 | Arginine--tRNA ligase | ArgS | 0.54 |
| Cell morphogenesis | ||||
| Q9RRJ4 | DR_2496 | UDP-N-acetylmuramoylalanine--D-glutamate ligase | MurD | 0.65 |
| Q9RWN8 | DR_0628 | UDP-N-acetylenolpyruvoylglucosamine reductase | MurB | 0.59 |
| Q9RXF1 | DR_0362 | D-alanine--D-alanine ligase | Ddl | 0.46 |
| Q9RWN9 | DR_0627 | UDP-N-acetylmuramate--L-alanine ligase | MurC | 0.42 |
| Q9RXL3 | DR_0297 | UDP-N-acetylmuramyl-tripeptide synthetase | MurE | 0.40 |
表3 与lon1有关的响应高温胁迫的差异表达蛋白
Table 3 Differentially expressed proteins associated with lon1 in response to high temperature stress
| UniProtKB | Locus | Predicted function | Protein name | FC |
|---|---|---|---|---|
| Protein fate(heat shock responsive chaperones and protease) | ||||
| Q9RV58 | DR_1172 | Chaperone-like protein | DosH | 13.37 |
| Q9RWQ9 | DR_0607 | 60 kD chaperone | GroL | 7.08 |
| Q9RVI3 | DR_1046 | Chaperone protein ClpB | ClpB | 4.70 |
| Q9RXG4 | DR_0349 | Lon protease | Lon2 | 4.52 |
| Q9RWR0 | DR_0606 | 10 kD chaperone | GroS | 4.46 |
| Q9RY24 | DR_0128 | Chaperone protein GrpE | GrpE | 3.24 |
| DNA metabolism(repair and recombination) | ||||
| Q9RXI7 | DR_0326 | DNA damage response protein D | DdrD | 2.59 |
| Q9RS64 | DR_2263 | DNA protection during starvation protein 1 | Dps1 | 2.88 |
| Q9RY51 | DR_0099 | Single-stranded DNA-binding protein | Ssb | 3.25 |
| Q9RSJ6 | DR_2128 | DNA-directed RNA polymerase subunit alpha | RpoA | 2.06 |
| Q9RRE4 | DR_2548 | Transcriptional regulatory protein | 7.75 | |
| Membrane functions and transport | ||||
| Q9RWU0 | DR_0575 | Protein translocase subunit SecA | SecA | 2.18 |
| P56867 | DR_2508 | Hexagonally packed intermediate-layer surface protein | Hpi | 3.48 |
| Q9RWH3 | DR_0695 | V-type ATP synthase subunit | AtpI | 1.57 |
| Translation | ||||
| Q9RST0 | DR_2043 | 50S ribosomal protein L7/L12 | RplL | 0.14 |
| Central intermediary metabolism(synthesis of nitrogen,sulfur,polyamine compounds) | ||||
| Q9RR70 | DR_2627 | Fumarate hydratase class II | FumC | 7.59 |
| Q9RWB2 | DR_0757 | Citrate synthase | GltA | 3.06 |
| Q9RZ06 | DR_A0147 | Histidine ammonia-lyase | HutH | 2.66 |
| Q9RTN7 | DR_1720 | Aconitate hydratase A | Acn | 2.10 |
| P56861 | DR_A0014 | Adenylyl-sulfate kinase | CysC | 0.23 |
| Amino acid biosynthesis and energy metabolism | ||||
| Q9RRT0 | DR_2405 | Demethylmenaquinone methyltransferase | MenG | 0.51 |
| Q9RV69 | DR_1161 | 5-hydroxyisourate hydrolase | 0.33 | |
| Q9RV98 | DR_1131 | Ferrochelatase | HemH | 2.04 |
| Q9RRC4 | DR_2568 | Arginine--tRNA ligase | ArgS | 0.54 |
| Cell morphogenesis | ||||
| Q9RRJ4 | DR_2496 | UDP-N-acetylmuramoylalanine--D-glutamate ligase | MurD | 0.65 |
| Q9RWN8 | DR_0628 | UDP-N-acetylenolpyruvoylglucosamine reductase | MurB | 0.59 |
| Q9RXF1 | DR_0362 | D-alanine--D-alanine ligase | Ddl | 0.46 |
| Q9RWN9 | DR_0627 | UDP-N-acetylmuramate--L-alanine ligase | MurC | 0.42 |
| Q9RXL3 | DR_0297 | UDP-N-acetylmuramyl-tripeptide synthetase | MurE | 0.40 |
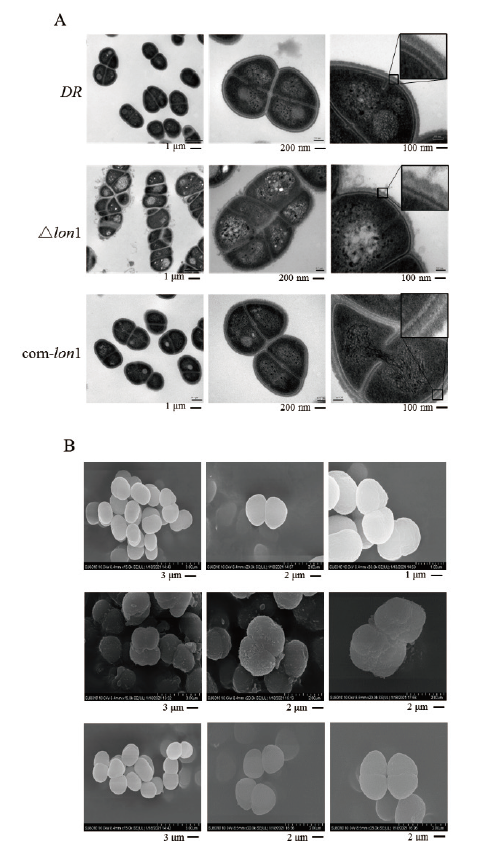
图6 耐辐射异常球菌野生型DR、△lon1以及comlon1的电镜图 A:透射电镜结果;B:扫描电镜结果
Fig. 6 Electron microscopy images of WT D. radiodurans DR,△lon1 and comlon1 A:Transmission electron microscopy(TEM). B:Scanning electron microscopy(SEM)
| [1] | Gur E, Ottofueling R, Dougan DA. Machines of destruction - AAA+ proteases and the adaptors that control them[M]// Subcellular Biochemistry. Dordrecht: Springer Netherlands, 2013:3-33. |
| [2] |
Mahmoud SA, Chien P. Regulated proteolysis in bacteria[J]. Annu Rev Biochem, 2018, 87:677-696.
doi: 10.1146/annurev-biochem-062917-012848 pmid: 29648875 |
| [3] | 刘郁夫, 董浩, 孙石静, 等. 细菌Lon蛋白酶研究进展[J]. 微生物学通报, 2019, 46(7):1706-1711. |
| Liu YF, Dong H, Sun SJ, et al. Research progress on bacterial Lon protease[J]. Microbiol China, 2019, 46(7):1706-1711. | |
| [4] | Gur E. The lon AAA+ protease[M]// Subcellular Biochemistry. Dordrecht: Springer Netherlands, 2013:35-51. |
| [5] |
Lin CC, Su SC, Su MY, et al. Structural insights into the allosteric operation of the lon AAA+ protease[J]. Structure, 2016, 24(5):667-675.
doi: 10.1016/j.str.2016.03.001 URL |
| [6] |
Tsilibaris V, Maenhaut-Michel G, van Melderen L. Biological roles of the Lon ATP-dependent protease[J]. Res Microbiol, 2006, 157(8):701-713.
pmid: 16854568 |
| [7] |
Jonas K, Liu J, Chien P, et al. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA[J]. Cell, 2013, 154(3):623-636.
doi: 10.1016/j.cell.2013.06.034 URL |
| [8] |
Adler HI, Hardigree AA. Cell elongation in strains of Escherichia coli[J]. J Bacteriol, 1964, 87(5):1240-1242.
doi: 10.1128/jb.87.5.1240-1242.1964 pmid: 5334970 |
| [9] |
Howard-Flanders P, Simson E, Theriot L. A locus that controls filament formation and sensitivity to radiation in Escherichia coli k-12[J]. Genetics, 1964, 49:237-246.
doi: 10.1093/genetics/49.2.237 URL |
| [10] |
Fernández L, Breidenstein EBM, Taylor PK, et al. Interconnection of post-transcriptional regulation:The RNA-binding protein Hfq is a novel target of the Lon protease in Pseudomonas aeruginosa[J]. Sci Rep, 2016, 6:26811.
doi: 10.1038/srep26811 pmid: 27229357 |
| [11] |
Kim H, Lee H, Shin D. Lon-mediated proteolysis of the FeoC protein prevents Salmonella enterica from accumulating the Fe(II)transporter FeoB under high-oxygen conditions[J]. J Bacteriol, 2015, 197(1):92-98.
doi: 10.1128/JB.01826-14 URL |
| [12] |
Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans[J]. Microbiol Mol Biol Rev, 2011, 75(1):133-191.
doi: 10.1128/MMBR.00015-10 URL |
| [13] |
Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans[J]. Nat Rev Microbiol, 2009, 7(3):237-245.
doi: 10.1038/nrmicro2073 URL |
| [14] |
Daly MJ, Gaidamakova EK, Matrosova VY, et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans[J]. PLoS One, 2010, 5(9):e12570.
doi: 10.1371/journal.pone.0012570 URL |
| [15] | 刘盈盈. 耐辐射异常球菌亲水蛋白DohL具有类分子伴侣和核酸内切酶功能并参与氧化胁迫保护[D]. 北京: 中国农业科学院, 2019. |
| Liu YY. Chaperone-like and catalytic functions in Deinococcus radiodurans of hydrophilic protein DohL involved in protective against oxidative stress[D]. Beijing: Chinese Academy of Agricultural Sciences, 2019. | |
| [16] | 陈晓楠. 耐辐射异常球菌黄嘌呤脱氢酶基因在氧化胁迫反应中的作用[D]. 北京: 中国农业科学院, 2020. |
| Chen XN. Role of xanthine dehydrogenase gene in oxidative stress response of Deinococcus radiodurans R1[D]. Beijing: Chinese Academy of Agricultural Sciences, 2020. | |
| [17] |
Karlin S, Mrazek J. Predicted highly expressed and putative alien genes of Deinococcus radiodurans and implications for resistance to ionizing radiation damage[J]. PNAS, 2001, 98(9):5240-5245.
pmid: 11296249 |
| [18] |
Heikkila JJ, Schultz GA, Iatrou K, et al. Expression of a set of fish genes following heat or metal ion exposure[J]. J Biol Chem, 1982, 257(20):12000-12005.
pmid: 7118927 |
| [19] |
Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control:folding, refolding, and degrading proteins[J]. Science, 1999, 286(5446):1888-1893.
pmid: 10583944 |
| [20] |
Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance[J]. Plant Biotechnol J, 2017, 15(4):405-414.
doi: 10.1111/pbi.12659 URL |
| [21] |
Su SC, Stephens BB, Alexandre G, et al. Lon protease of the alpha-proteobacterium Agrobacterium tumefaciens is required for normal growth, cellular morphology and full virulence[J]. Microbiol Read Engl, 2006, 152(Pt 4):1197-1207.
doi: 10.1099/mic.0.28657-0 URL |
| [22] |
Breidenstein EB, Janot L, Strehmel J, et al. The Lon protease is essential for full virulence in Pseudomonas aeruginosa[J]. PLoS One, 2012, 7(11):e49123.
doi: 10.1371/journal.pone.0049123 URL |
| [23] |
Jukič M, Gobec S, Sova M. Reaching toward underexplored targets in antibacterial drug design[J]. Drug Dev Res, 2019, 80(1):6-10.
doi: 10.1002/ddr.21465 URL |
| [24] |
Gottesman S, Halpern E, Trisler P. Role of SulA and sulB in filamentation by lon mutants of Escherichia coli K-12[J]. J Bacteriol, 1981, 148(1):265-273.
doi: 10.1128/jb.148.1.265-273.1981 pmid: 7026534 |
| [25] |
Mizusawa S, Gottesman S. Protein degradation in Escherichia coli:the lon gene controls the stability of SulA protein[J]. PNAS, 1983, 80(2):358-362.
pmid: 6300834 |
| [1] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [2] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [3] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [4] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [5] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [6] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [7] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [8] | 许睿, 祝英方. 中介体复合物在植物非生物胁迫应答中的功能[J]. 生物技术通报, 2023, 39(11): 54-60. |
| [9] | 孙雨桐, 刘德帅, 齐迅, 冯美, 黄栩筝, 姚文孔. 茉莉酸调控植物生长发育和胁迫的研究进展[J]. 生物技术通报, 2023, 39(11): 99-109. |
| [10] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [11] | 安昌, 陆琳, 沈梦千, 陈盛圳, 叶康卓, 秦源, 郑平. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16. |
| [12] | 位欣欣, 兰海燕. 植物MYB转录因子调控次生代谢及逆境响应的研究进展[J]. 生物技术通报, 2022, 38(8): 12-23. |
| [13] | 王慧, 马艺文, 乔正浩, 常彦彩, 术琨, 丁海萍, 聂永心, 潘光堂. AOX基因家族的结构和功能特征分析[J]. 生物技术通报, 2022, 38(7): 160-170. |
| [14] | 周国彦, 银珊珊, 高佳鑫, 武春成, 闫立英, 谢洋. 黄瓜AHP基因家族的鉴定及其非生物胁迫表达分析[J]. 生物技术通报, 2022, 38(6): 112-119. |
| [15] | 刘坤, 李国婧, 杨杞. 参与植物非生物逆境响应的DREB/CBF转录因子研究进展[J]. 生物技术通报, 2022, 38(5): 201-214. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||