








生物技术通报 ›› 2022, Vol. 38 ›› Issue (5): 74-83.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0981
雷君1,2( ), 陈勤1,2, 邓兵1,2, 张金渝3, 刘迪秋1,2, 崔秀明1,2, 葛锋1,2(
), 陈勤1,2, 邓兵1,2, 张金渝3, 刘迪秋1,2, 崔秀明1,2, 葛锋1,2( )
)
收稿日期:2021-08-02
出版日期:2022-05-26
发布日期:2022-06-10
作者简介:雷君,女,硕士研究生,研究方向:药用植物生物技术;E-mail: 基金资助:
LEI Jun1,2( ), CHEN Qin1,2, DENG Bing1,2, ZHANG Jin-yu3, LIU Di-qiu1,2, CUI Xiu-ming1,2, GE Feng1,2(
), CHEN Qin1,2, DENG Bing1,2, ZHANG Jin-yu3, LIU Di-qiu1,2, CUI Xiu-ming1,2, GE Feng1,2( )
)
Received:2021-08-02
Published:2022-05-26
Online:2022-06-10
摘要:
旨在明确PnMYB1转录因子对三七皂苷生物合成具有调控作用。利用RACE技术获得PnMYB1基因全长,对PnMYB1进行系统发育树分析;构建PnMYB1植物过表达载体并侵染三七细胞,检测转基因三七细胞中人参皂苷R1、Rg1、Re、Rb1和Rd的含量;将PnMYB1与鲨烯合酶(PnSS)、鲨烯环氧化酶(PnSE)、达玛烯二醇合成酶(PnDS)和环阿屯醇合成酶(PnCAS)等参与三七皂苷生物合成途径的关键酶的基因启动子共转染烟草叶片,进行瞬时表达分析,利用GUS表达系统验证PnMYB1转录因子能否与三七皂苷生物合成关键酶基因的启动子相互作用。结果显示,PnMYB1转录因子属于R2R3-MYB家族;在过表达PnMYB1的三七细胞中,五种重要三萜皂苷在转基因细胞中均有不同程度的增加,进一步分析证明PnMYB1转录因子通过激活PnSE和PnDS的启动子,促使PnSE和PnDS的表达水平显著升高,进而实现对三七皂苷生物合成的调控。PnMYB1转录因子可以同时调控三七皂苷生物合成途径中两个关键酶基因的表达,从而影响三七皂苷的生物合成。
雷君, 陈勤, 邓兵, 张金渝, 刘迪秋, 崔秀明, 葛锋. R2R3-MYB转录因子PnMYB1调控三七皂苷生物合成[J]. 生物技术通报, 2022, 38(5): 74-83.
LEI Jun, CHEN Qin, DENG Bing, ZHANG Jin-yu, LIU Di-qiu, CUI Xiu-ming, GE Feng. Biosynthesis of Panax notoginseng Saponins Regulated by R2R3-MYB Transcription Factor PnMYB1[J]. Biotechnology Bulletin, 2022, 38(5): 74-83.
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| PnGAPDH-F | CTTTGGTTTAAGGAACCCAGAGG |
| PnGAPDH-R | AAGGGGAGCAAGGCAGTTAGTAG |
| PnSS-F | CGAGCACTTGACACTGTTGAGGAT |
| PnSS-R | CTATTGCCTCCTGGTAACCGTTTC |
| PnDS-F | CAAGCACACGATGGTCACTGGC |
| PnDS-R | CATTTTGATGGTTGTAAACGAAGCG |
| PnSE-F | AGGTGAACTTCTACAACCAGGAGGC |
| PnSE-R | CTCAACCAGAGATGTAACAGTCCCC |
| PnCAS-F | GAAATTATACCCTGACCACCGT |
| PnCAS-R | CCAAACCTTTTACACCGAACC |
| PnMYB1-F | GCAGGTTTAAAGAGATGTGGGAAG |
| PnMYB1-R | CTGAAGCAGAGGAGGAGTGATTG |
表1 实时荧光定量PCR的引物
Table 1 Primers used for qRT-PCR
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| PnGAPDH-F | CTTTGGTTTAAGGAACCCAGAGG |
| PnGAPDH-R | AAGGGGAGCAAGGCAGTTAGTAG |
| PnSS-F | CGAGCACTTGACACTGTTGAGGAT |
| PnSS-R | CTATTGCCTCCTGGTAACCGTTTC |
| PnDS-F | CAAGCACACGATGGTCACTGGC |
| PnDS-R | CATTTTGATGGTTGTAAACGAAGCG |
| PnSE-F | AGGTGAACTTCTACAACCAGGAGGC |
| PnSE-R | CTCAACCAGAGATGTAACAGTCCCC |
| PnCAS-F | GAAATTATACCCTGACCACCGT |
| PnCAS-R | CCAAACCTTTTACACCGAACC |
| PnMYB1-F | GCAGGTTTAAAGAGATGTGGGAAG |
| PnMYB1-R | CTGAAGCAGAGGAGGAGTGATTG |
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| PnCAS-GSP1 | AGCTTTCTCGATGTCCGCAAGCTCTTC |
| PnCAS-GSP2 | GGATTTCCTCCCTCTGCAATTTTAAGC |
| PnDS-GSP1 | TCCCAAAATGCTTAAAGCCTCGATCCA |
| PnDS-GSP2 | CTTGTAGGGCTTATTGTTATGCAGATTGTG |
| PnSS-GSP1 | CATTTCATGTCCCGTTTTCCTGTAAGAAC |
| PnSS-GSP2 | ACAGTGTTTCTTTCCAGCGAGGACTCC |
| PnDSP-F | ATCGATTCAATACCGTGTGCTACTATGCAAC |
| PnDSP-R | GGATCCTGGTTATGTGGTGTACATAGATGGC |
| PnCASP-F | AAGCTTTGAGGGGCCAAATTCGTTG |
| PnCASP-R | TCTAGACACTCTGCACACAAATTTAGCTCC |
| PnSEP-F | AAGCTTTTGTGGGTCAGATCAGATGGA |
| PnSEP-R | GGATCCGGTGTTGGTTGGACGTTCAC |
表2 启动子克隆的引物
Table 2 Primers used for promoter clone
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| PnCAS-GSP1 | AGCTTTCTCGATGTCCGCAAGCTCTTC |
| PnCAS-GSP2 | GGATTTCCTCCCTCTGCAATTTTAAGC |
| PnDS-GSP1 | TCCCAAAATGCTTAAAGCCTCGATCCA |
| PnDS-GSP2 | CTTGTAGGGCTTATTGTTATGCAGATTGTG |
| PnSS-GSP1 | CATTTCATGTCCCGTTTTCCTGTAAGAAC |
| PnSS-GSP2 | ACAGTGTTTCTTTCCAGCGAGGACTCC |
| PnDSP-F | ATCGATTCAATACCGTGTGCTACTATGCAAC |
| PnDSP-R | GGATCCTGGTTATGTGGTGTACATAGATGGC |
| PnCASP-F | AAGCTTTGAGGGGCCAAATTCGTTG |
| PnCASP-R | TCTAGACACTCTGCACACAAATTTAGCTCC |
| PnSEP-F | AAGCTTTTGTGGGTCAGATCAGATGGA |
| PnSEP-R | GGATCCGGTGTTGGTTGGACGTTCAC |
| 人参皂苷标准品 Ginsenoside standards | 回归方程 Regression | R2 |
|---|---|---|
| Rg1 | y = 3944.9x +69.409 | 0.9997 |
| Re | y = 2646.8x+22.808 | 0.9999 |
| Rb1 | y = 3227.3x+49.436 | 0.9998 |
| Rd | y = 2953.5x+41.408 | 0.9998 |
| R1 | y = 2705.6x+50.697 | 0.9996 |
表3 五种重要单体皂苷线性回归方程
Table 3 Linear regression equations for five major mono-mer saponins
| 人参皂苷标准品 Ginsenoside standards | 回归方程 Regression | R2 |
|---|---|---|
| Rg1 | y = 3944.9x +69.409 | 0.9997 |
| Re | y = 2646.8x+22.808 | 0.9999 |
| Rb1 | y = 3227.3x+49.436 | 0.9998 |
| Rd | y = 2953.5x+41.408 | 0.9998 |
| R1 | y = 2705.6x+50.697 | 0.9996 |
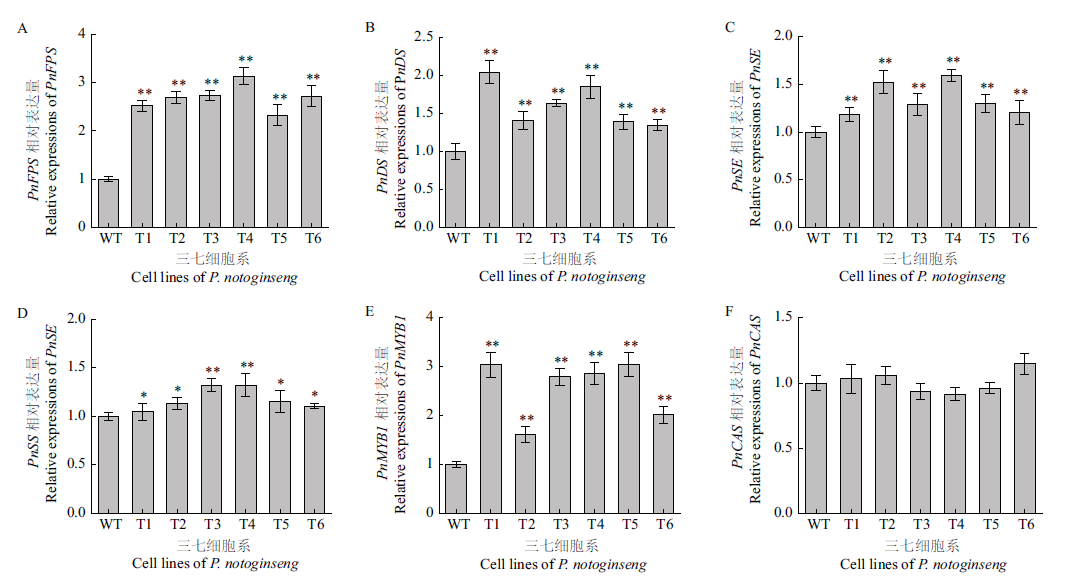
图3 三七细胞系T1-T6的PnFPS(A)、PnDS(B)、PnSE(C)、PnSS(D)、PnMYB1(E)和PnCAS(F)基因相对表达量 WT:野生型三七细胞;对照组比较:**P<0.01;*P<0.05. 下同
Fig. 3 Relative expressions of gene PnFPS(A),PnDS(B),PnSE(C),PnSS(D),PnMYB1(E)and PnCAS(F)in the T1-T6 cell lines of P. notoginseng WT:Wild type cell of P. notoginseng;**P<0.01 vs control group;*P<0.05 vs control group. The same below
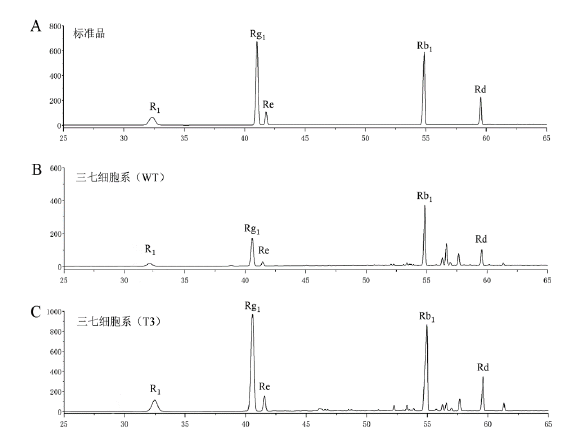
图5 转PnMYB1三七细胞系的HPLC分析 A:标准品;B:野生型三七细胞;C:转PnMYB1 T3三七细胞株系
Fig. 5 HPLC analysis of transformed PnMYB1 P. notogin-seng cell line A:Standard. B:Wild-type P. notoginseng cell. C:Transformed PnMYB1 T3 P. notoginseng cell line
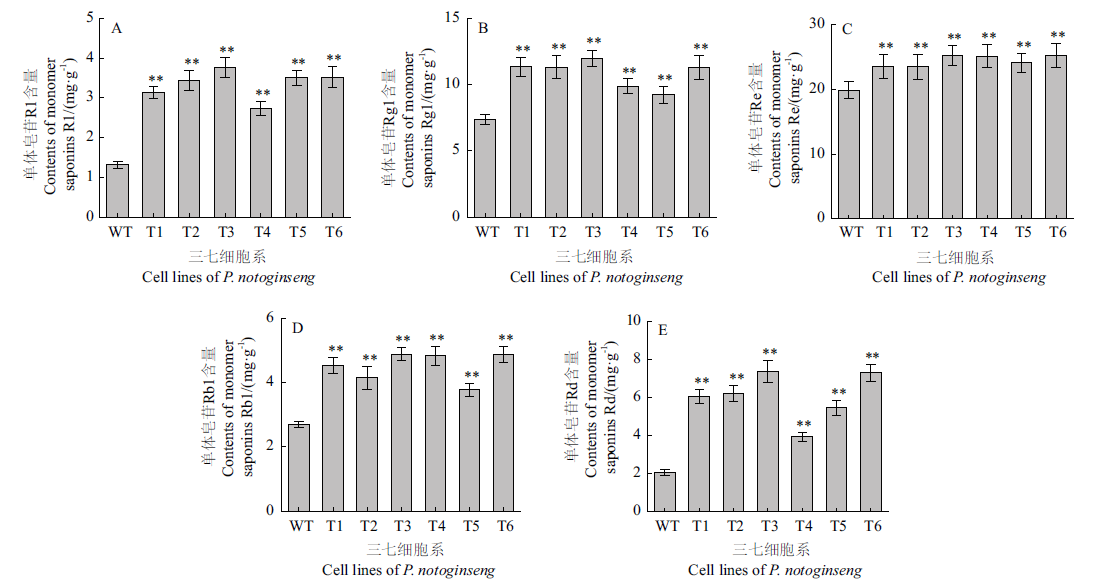
图6 三七细胞系T1-T6单体皂苷R1(A)、Rg1(B)、Re(C)、Rb1(D)和Rd(E)含量
Fig. 6 Contents of monomer saponins R1(A),Rg1(B),Re(C),Rb1(D)and Rd(E)in T1-T6 transgenic cell lines of P. notoginseng
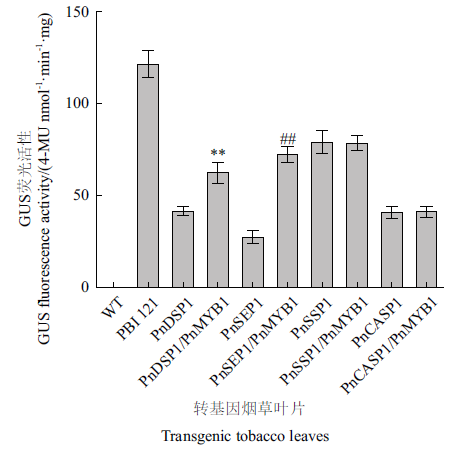
图7 GUS荧光活性分析 PnDSP1、PnSEP1、PnSSP1和PnCASP1分别表示对应的启动子片段单独转染的烟草叶片;PnDSP1/PnMYB1、PnSEP1/PnMYB1、PnSSP1/PnMYB1和PnCASP1/PnMYB1分别表示PnDSP1、PnSEP1、PnSSP1和PnCASP1启动子片段分别与PnMYB1共同转染的烟草叶片;PBI 121:PBI 121转染的野生型烟草叶片;WT:野生烟草叶片(与PnDSP1比较:**P<0.01,与PnSEP1比较:##P <0.01)
Fig. 7 GUS fluorescence activity analysis PnDSP1,PnSEP1,PnSSP1 and PnCASP1 refer to the tobacco leaves transgenic with PnDSP1,PnSEP1,PnSSP1 and PnCASP1 promoter fragments,respectively. PnSSP1/PnMYB1,PnSEP1/PnMYB1,PnDSP1/PnMYB1 and PnCASP1/PnMYB1 refers to the tobacco leaves that PnSSP1,PnSEP1,PnDSP1 and PnCASP1 promoter fragments co-transfected with PnMYB1,respectively. PBI 121:PBI 121-transfected wild-type tobacco leaves. WT:Wild tobacco leaves.(** P < 0.01 vs PnDSP1;and ## P <0.01 vs PnSEP1)
| [1] |
Ng TB. Pharmacological activity of Sanchi ginseng(Panax notoginseng)[J]. J Pharm Pharmacol, 2010, 58(8):1007-1019.
doi: 10.1211/jpp.58.8.0001 URL |
| [2] |
Kuzuyama T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units[J]. Biosci Biotechnol Biochem, 2002, 66(8):1619-1627.
doi: 10.1271/bbb.66.1619 URL |
| [3] |
Kim DH. Chemical Diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng[J]. J Ginseng Res, 2012, 36(1):1-15.
doi: 10.5142/jgr.2012.36.1.1 URL |
| [4] |
Zhang JZ. Overexpression analysis of plant transcription factors[J]. Curr Opin Plant Biol, 2003, 6(5):430-440.
doi: 10.1016/S1369-5266(03)00081-5 URL |
| [5] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants:a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5):735-747.
doi: 10.1093/molbev/msg088 URL |
| [6] |
Ma D, Pu G, Lei C, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the Amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis[J]. Plant Cell Physiol, 2009, 50(12):2146-2161.
doi: 10.1093/pcp/pcp149 URL |
| [7] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10):573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [8] |
Bedon F, Bomal C, Caron S, et al. Subgroup 4 R2R3-MYBs in conifer trees:gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses[J]. J Exp Bot, 2010, 61(14):3847-3864.
doi: 10.1093/jxb/erq196 URL |
| [9] | Li CY, Leopold AL, Sander GW, et al. CrBPF1 overexpression alters transcript levels of terpenoid indole alkaloid biosynthetic and regulatory genes[J]. Front Plant Sci, 2015, 6:818. |
| [10] | 徐俊雄, 覃建兵, 王岩岩, 等. 虎杖MYB转录因子PcMYB1的表达特性和功能研究[J]. 中草药, 2020, 51(3):726-732. |
| Xu JX, Qin JB, Wang YY, et al. Expression and function analysis of transcription factor PcMYB1 from Polygonum cuspidatum[J]. Chin Tradit Herb Drugs, 2020, 51(3):726-732. | |
| [11] | 应宇翔, 何凤明, 张云峰, 等. 灯盏花MYB基因克隆及其荧光表达载体的构建[J]. 中草药, 2017, 48(20):4306-4315. |
| Ying YX, He FM, Zhang YF, et al. Cloning of MYB gene and construction of greenfluorescent protein expression vector in Erigeron breviscapus[J]. Chin Tradit Herb Drugs, 2017, 48(20):4306-4315. | |
| [12] |
Puissant C, Houdebine LM. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction[J]. BioTechniques, 1990, 8(2):148-149.
pmid: 1690559 |
| [13] |
Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction[J]. Anal Biochem, 1987, 162(1):156-159.
doi: 10.1006/abio.1987.9999 pmid: 2440339 |
| [14] |
Dai Y, Qin Q, Dai D, et al. Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata[J]. Biotechnol Lett, 2009, 31(11):1801-1809.
doi: 10.1007/s10529-009-0068-4 URL |
| [15] |
Höfgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation[J]. Nucleic Acids Res, 1988, 16(20):9877.
pmid: 3186459 |
| [16] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [17] |
Wako T, Kimura S, Chikagawa Y, et al. Characterization of MYB proteins acting as transcriptional regulatory factors for carrot phenylalanine ammonia-lyase gene(DcPAL3)[J]. Plant Biotechnol, 2010, 27(2):131-139.
doi: 10.5511/plantbiotechnology.27.131 URL |
| [18] |
Wang N, Xu H, Jiang S, et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple(Malus sieversii f. niedzwetzkyana)[J]. Plant J, 2017, 90(2):276-292.
doi: 10.1111/tpj.13487 URL |
| [19] |
Gonzalez A, Zhao M, Leavitt JM, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings[J]. Plant J, 2008, 53(5):814-827.
doi: 10.1111/j.1365-313X.2007.03373.x URL |
| [20] |
Dai X, Xu Y, Ma Q, et al. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis[J]. Plant Physiol, 2007, 143(4):1739-1751.
doi: 10.1104/pp.106.094532 URL |
| [21] |
Elomaa P, Uimari A, Mehto M, et al. Activation of anthocyanin biosynthesis in Gerbera hybrida(Asteraceae)suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots[J]. Plant Physiol, 2003, 133(4):1831-1842.
pmid: 14605235 |
| [22] |
Deluc L, Bogs J, Walker AR, et al. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries[J]. Plant Physiol, 2008, 147(4):2041-2053.
doi: 10.1104/pp.108.118919 URL |
| [23] |
Li CH, Qiu J, Yang GS, et al. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum(Hort. )[J]. Plant Cell Rep, 2016, 35(10):2151-2165.
doi: 10.1007/s00299-016-2025-8 URL |
| [24] |
Lin WK, Bolitho K, Grafton K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae[J]. BMC Plant Biol, 2010, 10:50.
doi: 10.1186/1471-2229-10-50 pmid: 20302676 |
| [25] |
Lee MH, Jeong JH, Seo JW, et al. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene[J]. Plant Cell Physiol, 2004, 45(8):976-984.
doi: 10.1093/pcp/pch126 URL |
| [26] |
Deng B, Zhang P, Ge F, et al. Enhancement of triterpenoid saponins biosynthesis in Panax notoginseng cells by co-overexpressions of 3-hydroxy-3-methylglutaryl CoA reductase and squalene synthase genes[J]. Biochem Eng J, 2017, 122:38-46.
doi: 10.1016/j.bej.2017.03.001 URL |
| [27] |
Laitinen RA, Ainasoja M, Broholm SK, et al. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida[J]. J Exp Bot, 2008, 59(13):3691-3703.
doi: 10.1093/jxb/ern216 pmid: 18725377 |
| [28] |
Leung KW, Wong AS. Pharmacology of ginsenosides:a literature review[J]. Chin Med, 2010, 5:20.
doi: 10.1186/1749-8546-5-20 URL |
| [29] |
Lee JJ, Kwon HK, Jung IH, et al. Anti-cancer activities of ginseng extract fermented with Phellinus linteus[J]. Mycobiology, 2009, 37(1):21-27.
doi: 10.4489/MYCO.2009.37.1.021 URL |
| [30] |
Ni N, Liu Q, Ren H, et al. Ginsenoside Rb1 protects rat neural progenitor cells against oxidative injury[J]. Molecules, 2014, 19(3):3012-3024.
doi: 10.3390/molecules19033012 URL |
| [31] | 陈勤, 雷君, 葛锋, 等. 人参属三萜皂苷骨架修饰的研究进展[J]. 中药材, 2020, 11:2831-2837. |
| Chen Q, Lei J, Ge F, et al. The research progress of ginseng triterpenoid saponin skeleton modifying enzyme[J]. Journal of Chinese Medicinal Materials, 2020, 11:2831-2837. |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [7] | 赵林艳, 徐武美, 王豪吉, 王昆艳, 魏富刚, 杨绍周, 官会林. 施用生物炭对连作三七根际真菌群落与存活率的影响[J]. 生物技术通报, 2023, 39(7): 219-227. |
| [8] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [9] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [10] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [11] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [12] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [13] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| [14] | 赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125. |
| [15] | 孙海航, 官会林, 王旭, 王童, 李泓霖, 彭文洁, 刘柏桢, 樊芳玲. 生物炭对三七连作土壤性质及真菌群落的影响[J]. 生物技术通报, 2023, 39(2): 221-231. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||