








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 304-310.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0387
赵昕1,2( ), 杜玉瑶1,2, 殷子扬1,2, 毛淑红1,2(
), 杜玉瑶1,2, 殷子扬1,2, 毛淑红1,2( )
)
收稿日期:2023-04-22
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
毛淑红,女,博士,教授,研究方向:甾体药物微生物转化;E-mail: shuhongmao@tust. edu.cn作者简介:赵昕,女,硕士,研究方向:生物催化与生物转化;E-mail: 18332862765@163.com
基金资助:
ZHAO Xin1,2( ), DU Yu-yao1,2, YIN Zi-yang1,2, MAO Shu-hong1,2(
), DU Yu-yao1,2, YIN Zi-yang1,2, MAO Shu-hong1,2( )
)
Received:2023-04-22
Published:2023-10-26
Online:2023-11-28
摘要:
胆固醇7α-羟化酶(CYP7A1)是胆固醇分解为胆汁酸的限速酶,其催化胆固醇为7α-羟基胆固醇,7α-羟基胆固醇是重要的甾体药物及中间体。以人源CYP7A1作为研究对象,首先构建了重组毕赤酵母/pPIC3.5K-CYP7A1;然后对7α-羟化酶进行分子改造,之后将酶与不同来源NADPH细胞色素氧化还原酶(CPR)适配,以提高目的产物7α-羟基胆固醇的产量。其中突变体G485A将7α-羟基胆固醇的产量提高了8.70%;5种不同来源的CPR与CYP7A1共表达毕赤酵母菌株均可提高7α-羟基胆固醇的产量,并且罗汉果(Siraitia grosvenorii)来源的CPR(SgCPR)与CYP7A1适配性最好,使7α-羟基胆固醇的产率提高了82.26%;在以上研究的基础上,构建突变体G485A与SgCPR共表达毕赤酵母菌株,最终7α-羟基胆固醇的产量提高了133.79%,其产量为0.25 mg/L。实验在毕赤酵母中成功异源表达人源胆固醇7α-羟化酶,并通过分子改造及与不同来源CPR适配的方法提高了7α-羟基胆固醇的产量。
赵昕, 杜玉瑶, 殷子扬, 毛淑红. 胆固醇7α-羟化酶在毕赤酵母中的异源表达[J]. 生物技术通报, 2023, 39(10): 304-310.
ZHAO Xin, DU Yu-yao, YIN Zi-yang, MAO Shu-hong. Allogeneic Expression of Cholesterol 7α-hydroxylase in Pichia pastoris[J]. Biotechnology Bulletin, 2023, 39(10): 304-310.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| CYP7A1-F | CGCGGATCCATGATGACCACCTCTCTGATT |
| CYP7A1-R | CCGGAATTCTTACAGATGTTTAAATTTATATTTAAATTCAAT |
| H111A-F | AAAGCCTTTGGTGCTAGGTCTATTGATCCAATGGATGGTAATAC |
| H111A-R | ACCAAAGGCTTTGGCAGAGGT |
| I114A-F | GGTCATAGGTCTGCTGATCCAATGGATGGTAATACCACTGA |
| I114A-R | AGACCTATGACCAAAGGCTTTGGC |
| V281A-F | AAAACCCATCTGGCTGTTCTGTGGGCCTCTCAAGCC |
| V281A-R | CAGATGGGTTTTGGCTTTTTCC |
| W284A-F | CTGGTTGTTCTGGCYGCCTCTCAAGCCAATACCATTCC |
| W284A-R | CAGAACAACCAGATGGGTTTTGG |
| G485A-F | CAGTCTAGAGCTGCTCTGGGTATTCTGCCACCACTGA |
| G485A-R | AGCTCTAGACTGATCCAGTGGTGGAC |
表1 CYP7A1基因野生型及突变型引物
Table 1 Wild-type and mutant primers of CYP7A1 gene
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| CYP7A1-F | CGCGGATCCATGATGACCACCTCTCTGATT |
| CYP7A1-R | CCGGAATTCTTACAGATGTTTAAATTTATATTTAAATTCAAT |
| H111A-F | AAAGCCTTTGGTGCTAGGTCTATTGATCCAATGGATGGTAATAC |
| H111A-R | ACCAAAGGCTTTGGCAGAGGT |
| I114A-F | GGTCATAGGTCTGCTGATCCAATGGATGGTAATACCACTGA |
| I114A-R | AGACCTATGACCAAAGGCTTTGGC |
| V281A-F | AAAACCCATCTGGCTGTTCTGTGGGCCTCTCAAGCC |
| V281A-R | CAGATGGGTTTTGGCTTTTTCC |
| W284A-F | CTGGTTGTTCTGGCYGCCTCTCAAGCCAATACCATTCC |
| W284A-R | CAGAACAACCAGATGGGTTTTGG |
| G485A-F | CAGTCTAGAGCTGCTCTGGGTATTCTGCCACCACTGA |
| G485A-R | AGCTCTAGACTGATCCAGTGGTGGAC |
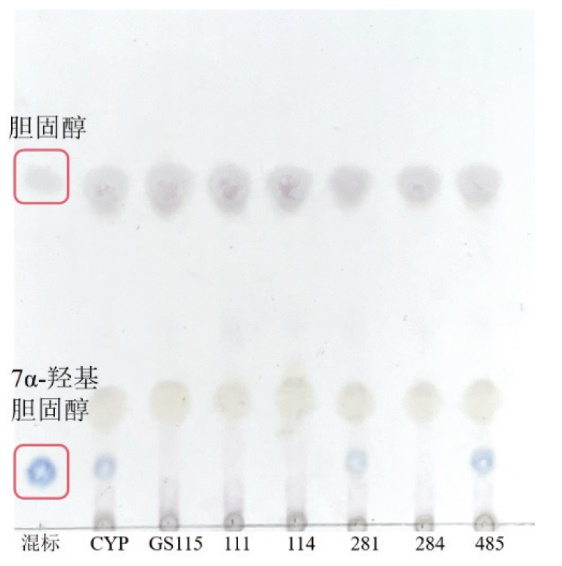
图2 重组毕赤酵母催化胆固醇TLC结果图 混标:胆固醇和7α-羟基胆固醇的混合标品;CYP:CYP7A1(WT);GS115:毕赤酵母GS115;111:突变体H111A;114:突变体I114A;281:突变体V281A;284:突变体W284A;485:突变体G485A
Fig. 2 TLC analysis of biotransformation result of cholesterol catalyzed by recombinant P. pastoris Mixed labels: Mixed standards of cholesterol and 7α-hydroxycholesterol; CYP: CYP7A1(WT); GS115: P. pastoris GS115; 111: mutant H111A; 114: mutant I114A; 281: mutant V281A; 284: mutant W284A; 485: mutant G485A
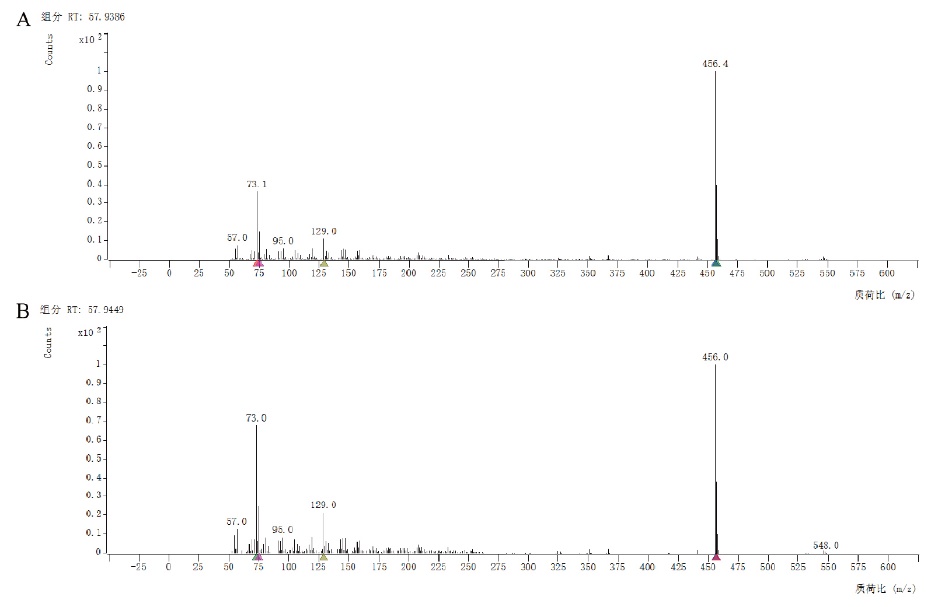
图3 重组毕赤酵母催化生成7α-羟基胆固醇的GC-MS分析图 A:7α-羟基胆固醇质谱图;B:重组毕赤酵母转化产物质谱图
Fig. 3 GC-MS analysis of the 7α-hydroxycholesterol catalyzed by recombinant P. pastoris A: Mass spectra of 7α-hydroxycholesterol. B: Mass spectra of recombinant P. pastoris products
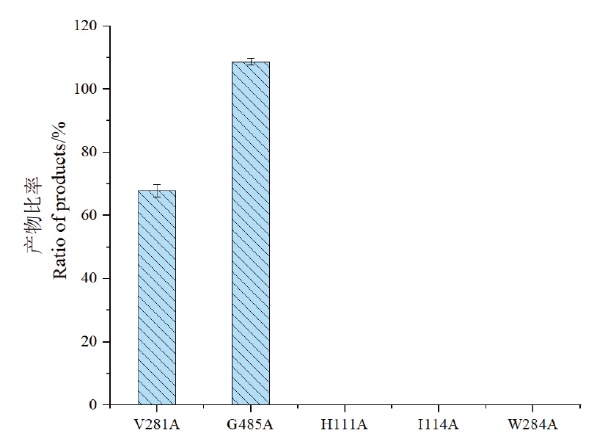
图4 突变毕赤酵母产率比较图 突变体H111A、I114A、V281A、W284A和G485A与CYP7A1(WT)重组毕赤酵母的产物比率
Fig. 4 Production abilities of the mutants when compared with the wild type Product ratio of mutant H111A, I114A, V281A, W284A and G485A to CYP7A1(WT)recombinant P. pastoris

图5 CYP7A1与不同来源CPR共表达毕赤酵母对胆固醇转化能力的影响 CYP7A1(WT)与ScCPR、hCPR、AnCPR、RatCPR和SgCPR共表达毕赤酵母,及G485A与SgCPR共表达毕赤酵母产物增长率(与CYP7A1(WT)重组毕赤酵母相比)
Fig. 5 Effects of P. pastoris co-expressed with CYP7A1 and CPR from different sources on cholesterol conversionThe growth rate of products of P. pastoris co-expressed with CYP7A1(WT)and CPR(ScCPR, hCPR, AnCPR, RatCPR and SgCPR)and P. pastoris co-expressed with G485A and SgCPR when compared with that of recombinant P. pastoris containing only CYP7A1(WT)

图6 CYP7A1与胆固醇分子对接结果图 黄色部分分子为胆固醇,其他部分为CYP7A1,蓝色标注位点为突变位点
Fig. 6 Docking results of CYP7A1 and cholesterol molecules The yellow molecule is cholesterol, the others are CYP7A1, and the blue-labeled sites are mutation sites
| [1] |
Qayyum F, Lauridsen BK, Frikke-Schmidt R, et al. Genetic variants in CYP7A1 and risk of myocardial infarction and symptomatic gallstone disease[J]. Eur Heart J, 2018, 39(22): 2106-2116.
doi: 10.1093/eurheartj/ehy068 pmid: 29529257 |
| [2] |
Rizzolo D, Kong B, Taylor RE, et al. Bile acid homeostasis in female mice deficient in Cyp7a1 and Cyp27a1[J]. Acta Pharm Sin B, 2021, 11(12): 3847-3856.
doi: 10.1016/j.apsb.2021.05.023 pmid: 35024311 |
| [3] |
Jiao JY, Zhu XJ, Zhou C, et al. Research progress on the immune microenvironment of the gallbladder in patients with cholesterol gallstones[J]. World J Gastrointest Surg, 2022, 14(9): 887-895.
doi: 10.4240/wjgs.v14.i9.887 URL |
| [4] |
Srivastava A, Choudhuri G, Mittal B. CYP7A1(-204 A>C; rs3808607 and-469 T>C; rs3824260)promoter polymorphisms and risk of gallbladder cancer in North Indian population[J]. Metabolism, 2010, 59(6): 767-773.
doi: 10.1016/j.metabol.2009.09.021 URL |
| [5] |
Omura T, Gotoh O. Evolutionary origin of mitochondrial cytochrome P450[J]. J Biochem, 2017, 161(5): 399-407.
doi: 10.1093/jb/mvx011 pmid: 28338801 |
| [6] |
Nowrouzi B, Rios-Solis L. Redox metabolism for improving whole-cell P450-catalysed terpenoid biosynthesis[J]. Crit Rev Biotechnol, 2022, 42(8): 1213-1237.
doi: 10.1080/07388551.2021.1990210 URL |
| [7] |
Li SY, Du L, Bernhardt R. Redox partners: function modulators of bacterial P450 enzymes[J]. Trends Microbiol, 2020, 28(6): 445-454.
doi: S0966-842X(20)30048-2 pmid: 32396826 |
| [8] |
Jiang YY, Li SY. Catalytic function and application of cytochrome P450 enzymes in biosynthesis and organic synthesis[J]. Chin J Org Chem, 2018, 38(9): 2307.
doi: 10.6023/cjoc201805055 URL |
| [9] |
Yasuda K, Sugimoto H, Hayashi K, et al. Protein engineering of CYP105s for their industrial uses[J]. Biochim Biophys Acta Proteins Proteom, 2018, 1866(1): 23-31.
doi: 10.1016/j.bbapap.2017.05.014 URL |
| [10] | 曾祥锐. 铁-卟啉氧化还原酶体系结构与氧化还原机理分析[J]. 化学工程与装备, 2017(1): 19-21. |
| Zeng XR. Analysis on the system structure and redox mechanism of iron-porphyrin oxidoreductase[J]. Chem Eng Equip, 2017(1): 19-21. | |
| [11] | Gillard J, Clerbaux LA, Nachit M, et al. Bile acids contribute to the development of non-alcoholic steatohepatitis in mice[J]. JHEP Rep, 2021, 4(1): 100387. |
| [12] |
Narasimhulu CA, Selvarajan K, Litvinov D, et al. Anti-atherosclerotic and anti-inflammatory actions of sesame oil[J]. J Med Food, 2015, 18(1): 11-20.
doi: 10.1089/jmf.2014.0138 pmid: 25562618 |
| [13] |
Slominski AT, Zmijewski MA, Semak I, et al. Cytochromes p450 and skin cancer: role of local endocrine pathways[J]. Anticancer Agents Med Chem, 2014, 14(1): 77-96.
doi: 10.2174/18715206113139990308 URL |
| [14] |
Karam WG, Chiang JY. Expression and purification of human cholesterol 7 alpha-hydroxylase in Escherichia coli[J]. J Lipid Res, 1994, 35(7): 1222-1231.
pmid: 7964183 |
| [15] |
Guengerich FP, Martin MV. Purification of cytochromes P450: products of bacterial recombinant expression systems[J]. Methods Mol Biol, 2006, 320: 31-38.
pmid: 16719372 |
| [16] |
Tempel W, Grabovec I, MacKenzie F, et al. Structural characterization of human cholesterol 7α-hydroxylase[J]. J Lipid Res, 2014, 55(9): 1925-1932.
doi: 10.1194/jlr.M050765 pmid: 24927729 |
| [17] |
Baghban R, Farajnia S, Rajabibazl M, et al. Yeast expression systems: overview and recent advances[J]. Mol Biotechnol, 2019, 61(5): 365-384.
doi: 10.1007/s12033-019-00164-8 pmid: 30805909 |
| [18] |
Mast N, Graham SE, Andersson U, et al. Cholesterol binding to cytochrome P450 7A1, a key enzyme in bile acid biosynthesis[J]. Biochemistry, 2005, 44(9): 3259-3271.
pmid: 15736936 |
| [19] |
Hlavica P, Schulze J, Lewis DFV. Functional interaction of cytochrome P450 with its redox partners: a critical assessment and update of the topology of predicted contact regions[J]. J Inorg Biochem, 2003, 96(2/3): 279-297.
doi: 10.1016/S0162-0134(03)00152-1 URL |
| [20] |
Khatri Y, Schifrin A, Bernhardt R. Investigating the effect of available redox protein ratios for the conversion of a steroid by a myxobacterial CYP260A1[J]. FEBS Lett, 2017, 591(8): 1126-1140.
doi: 10.1002/1873-3468.12619 pmid: 28281299 |
| [1] | 赵思佳, 王晓璐, 孙纪录, 田健, 张杰. 代谢工程改造毕赤酵母生产赤藓糖醇[J]. 生物技术通报, 2023, 39(8): 137-147. |
| [2] | 董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324. |
| [3] | 王玥, 高庆华, 董聪, 罗同阳, 王庆庆. 密码子优化的吡喃糖氧化酶基因在毕赤酵母中的表达[J]. 生物技术通报, 2022, 38(4): 269-277. |
| [4] | 杨威, 伍茜, 程建国, 罗燕, 王印, 杨泽晓, 姚学萍. 林麝干扰素α基因克隆、表达及转录调控分析[J]. 生物技术通报, 2022, 38(1): 194-204. |
| [5] | 廖兆民, 蔡俊, 林建国, 杜馨, 王常高. 黑曲霉葡萄糖氧化酶基因在毕赤酵母中的表达及产酶条件的优化[J]. 生物技术通报, 2021, 37(6): 97-107. |
| [6] | 杨悦, 陶妍, 谢晶, 钱韻芳. 基于重组毕赤酵母的草鱼C型溶菌酶生物合成及其抑菌活性[J]. 生物技术通报, 2021, 37(12): 169-179. |
| [7] | 熊亮斌, 孙吉, 刘显舟, 屈占国, 计雨情, 徐一新, 王风清. 分枝杆菌转化甾醇过程的中心代谢关键基因转录差异分析[J]. 生物技术通报, 2021, 37(10): 120-127. |
| [8] | 赵震, 王莎莎, 吕星星, 陶妍, 谢晶, 钱韻芳. 重组毕赤酵母产青蛤Mytimacin抗菌肽的表达研究[J]. 生物技术通报, 2020, 36(5): 150-158. |
| [9] | 闵琪, 高子涵, 姚银, 张华山, 熊海容, 张莉. 共表达HAC1和分子伴侣基因对甘露聚糖酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2020, 36(5): 159-168. |
| [10] | 李雅楠, 余利红, 陈新美, 杨浩萌, 黄火清. 来源于Penicillium sp.C1的水产用中性植酸酶基因在毕赤酵母中的表达及性质研究[J]. 生物技术通报, 2020, 36(2): 134-141. |
| [11] | 黄元霞, 彭传海, 丁宁, 邱忠平, 李星, 邹美慧. 一种三联乳酸菌的体外降胆固醇和抗氧化能力研究[J]. 生物技术通报, 2020, 36(12): 113-120. |
| [12] | 张亚莉, 陶妍, 谢晶, 钱韻芳. 厚壳贻贝Mytilin-1成熟肽在毕赤酵母中的重组表达及其抑菌活性[J]. 生物技术通报, 2019, 35(7): 54-60. |
| [13] | 侯成林, 杨艳坤, 陈嘉荔, 白仲虎. Mxr1磷酸化水平受Ptp调控机理的初步研究[J]. 生物技术通报, 2019, 35(7): 108-113. |
| [14] | 董聪, 高庆华, 王玥, 罗同阳. 基于密码子优化的FAD依赖葡萄糖脱氢酶在毕赤酵母中的高效表达及酶学性质[J]. 生物技术通报, 2019, 35(7): 114-120. |
| [15] | 刘进兰, 杨雪, 李双双, 张玉明, 柳峰松, 唐婷, 李红权. 家蝇抗菌肽Domesticin在毕赤酵母中的表达及抑菌活性检测[J]. 生物技术通报, 2019, 35(2): 109-115. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||