








生物技术通报 ›› 2023, Vol. 39 ›› Issue (2): 107-115.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0568
庞强强1,2,3( ), 孙晓东1,2,3(
), 孙晓东1,2,3( ), 周曼1,2,3, 蔡兴来1,2,3, 张文1,2,3, 王亚强1,2,3
), 周曼1,2,3, 蔡兴来1,2,3, 张文1,2,3, 王亚强1,2,3
收稿日期:2022-05-09
出版日期:2023-02-26
发布日期:2023-03-07
作者简介:庞强强,男,硕士,助理研究员,研究方向:菜心栽培与育种;E-mail: 基金资助:
PANG Qiang-qiang1,2,3( ), SUN Xiao-dong1,2,3(
), SUN Xiao-dong1,2,3( ), ZHOU Man1,2,3, CAI Xing-lai1,2,3, ZHANG Wen1,2,3, WANG Ya-qiang1,2,3
), ZHOU Man1,2,3, CAI Xing-lai1,2,3, ZHANG Wen1,2,3, WANG Ya-qiang1,2,3
Received:2022-05-09
Published:2023-02-26
Online:2023-03-07
摘要:
为探索菜心对高温胁迫的响应机制,发掘耐热相关基因,本研究采用全基因合成法(PAS)对前期从菜心高温胁迫转录组数据库中筛选到的1个差异表达基因进行克隆,并开展相关生物信息学分析,利用实时荧光定量PCR技术(RT-qPCR)分析该基因在高温胁迫下的表达模式。结果表明,克隆得到开放阅读框(ORF)为906 bp的基因,命名为BrHsfA3。该基因编码301个氨基酸,蛋白相对分子量为34.12 kD,理论等电点为5.00,为无信号肽的亲水性蛋白,存在丝氨酸、苏氨酸和酪氨酸3个磷酸化位点。BrHsfA3蛋白含有1个Hsf保守结构域,具有典型的DNA结合结构域(DBD)、寡聚化结构域(OD)、核定位信号结构域(NLS)和转录激活结构域(AHA),蛋白二级结构以α-螺旋和无规则卷曲为主。进化分析发现,BrHsfA3基因与甘蓝型油菜的亲缘关系最近。RT-qPCR分析结果显示,BrHsfA3在耐热自交系CX1-7叶、花中受高温诱导后表达量大幅上调,且显著高于热敏自交系CX7-3;根中表达量则在热敏自交系CX7-3中较高,在高温诱导后显著上调。本研究结果为进一步探讨BrHsfA3基因在菜心响应高温胁迫应答中的调控作用奠定基础。
庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115.
PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress[J]. Biotechnology Bulletin, 2023, 39(2): 107-115.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物用途Primer purpose |
|---|---|---|
| BrHsfA3-F1 | CGCCAGAATCATACTCCCGC | 基因合成 Gene synthesis |
| BrHsfA3-R1 | CCTTAGAGGACAGAAGCATC | 基因合成 Gene synthesis |
| BrHsfA3-F2 | GATGTGCTGCAAGGCGATTA | 基因合成 Gene synthesis |
| BrHsfA3-R2 | TTATGCTTCCGGCTCGTATG | 基因合成 Gene synthesis |
| BrActin-F | AGCAACTGGGATGACATGGA | RT-qPCR内参基因 RT-qPCR reference genes |
| BrActin-R | TCACCAGAGTCGAGCACAAT | RT-qPCR内参基因RT-qPCR reference genes |
| Q-BrHsfA3-F | GTCGACATAACAGACGTGGC | RT-qPCR |
| Q-BrHsfA3-R | CTGTCTGACGAAGCTGGAGA | RT-qPCR |
表1 基因克隆和表达分析引物
Table 1 Primers used for gene cloning and expression analysis
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物用途Primer purpose |
|---|---|---|
| BrHsfA3-F1 | CGCCAGAATCATACTCCCGC | 基因合成 Gene synthesis |
| BrHsfA3-R1 | CCTTAGAGGACAGAAGCATC | 基因合成 Gene synthesis |
| BrHsfA3-F2 | GATGTGCTGCAAGGCGATTA | 基因合成 Gene synthesis |
| BrHsfA3-R2 | TTATGCTTCCGGCTCGTATG | 基因合成 Gene synthesis |
| BrActin-F | AGCAACTGGGATGACATGGA | RT-qPCR内参基因 RT-qPCR reference genes |
| BrActin-R | TCACCAGAGTCGAGCACAAT | RT-qPCR内参基因RT-qPCR reference genes |
| Q-BrHsfA3-F | GTCGACATAACAGACGTGGC | RT-qPCR |
| Q-BrHsfA3-R | CTGTCTGACGAAGCTGGAGA | RT-qPCR |
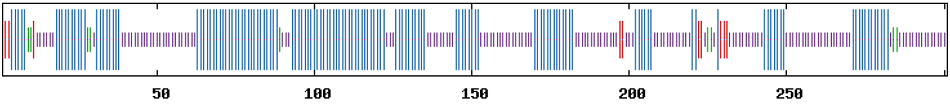
图6 BrHsfA3蛋白二级结构预测 横轴表示氨基酸位置;蓝色区域表示α-螺旋;紫色区域表示无规则卷曲;红色区域表示延伸链;绿色区域表示β-转角
Fig. 6 Prediction of BrHsfA3 protein secondary structure Horizontal axis indicates amino acid position. The blue part indicates alpha helix. The purple indicates random coil. The red part indicates extended strand. The green part indicates beta turn

图9 BrHsfA3基因在高温胁迫处理下的FPKM值 不同小写字母表示同一类型自交系植株差异显著(P< 0.05),下同
Fig. 9 FPKM value of BrHsfA3 gene under heat stress Different lowercase letters of the same inbred indicate significant difference(P<0.05),the same below
| [1] | 陈汉才, 吴增祥, 林悦欣, 等. 广东菜心、芥蓝研究现状与展望[J]. 广东农业科学, 2021, 48(9):62-71. |
| Chen HC, Wu ZX, Lin YX, et al. Research status and prospect of flowering Chinese cabbage and Chinese kale in Guangdong[J]. Guangdong Agric Sci, 2021, 48(9):62-71. | |
| [2] | 刘畅. 高温涝渍对菜心农艺性状和生理特性影响的研究[D]. 广州: 广州大学, 2020. |
| Liu C. Effect of high temperature and waterlogging on agronomic and physiological traits in flowering Chinese cabbage[D]. Guangzhou: Guangzhou University, 2020. | |
| [3] | 陈连珠, 张雪彬, 杨小锋. 根际高温对菜心生长及光合生理的影响[J]. 北方园艺, 2020(14):50-55. |
| Chen LZ, Zhang XB, Yang XF. Effects of rhizosphere high temperature on growth and photosynthetic physiology of Chinese flowering cabbages[J]. North Hortic, 2020(14):50-55. | |
| [4] |
Yue YZ, Jiang HY, Du JH, et al. Variations in physiological response and expression profiles of proline metabolism-related genes and heat shock transcription factor genes in Petunia subjected to heat stress[J]. Sci Hortic, 2019, 258:108811.
doi: 10.1016/j.scienta.2019.108811 URL |
| [5] | 司修洋, 梁文杰, 罗澜, 等. 甜瓜热激转录因子(Hsf)基因家族鉴定及生物信息学分析[J]. 中国蔬菜, 2020(11):23-32. |
| Si XY, Liang WJ, Luo L, et al. Identification and bioinformatics analysis of heat shock transcription factor(Hsf)gene family in melon[J]. China Veg, 2020(11):23-32. | |
| [6] |
Huang B, Huang ZN, Ma RF, et al. Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo(Phyllostachys edulis)[J]. Sci Rep, 2021, 11(1):16492.
doi: 10.1038/s41598-021-95899-3 pmid: 34389742 |
| [7] |
Shen CW, Yuan JP. Genome-wide characterization and expression analysis of the heat shock transcription factor family in pumpkin(Cucurbita moschata)[J]. BMC Plant Biol, 2020, 20:471.
doi: 10.1186/s12870-020-02683-y URL |
| [8] |
Li MY, Xie FJ, Li YW, et al. Genome-wide analysis of the heat shock transcription factor gene family in Brassica juncea:structure, evolution, and expression profiles[J]. DNA Cell Biol, 2020, 39(11):1990-2004.
doi: 10.1089/dna.2020.5922 URL |
| [9] |
Zhang Q, Geng J, Du YL, et al. Heat shock transcription factor(Hsf)gene family in common bean(Phaseolus vulgaris):genome-wide identification, phylogeny, evolutionary expansion and expression analyses at the sprout stage under abiotic stress[J]. BMC Plant Biol, 2022, 22(1):33.
doi: 10.1186/s12870-021-03417-4 pmid: 35031009 |
| [10] |
Zhou M, Zheng SG, Liu R, et al. Genome-wide identification, phylogenetic and expression analysis of the heat shock transcription factor family in bread wheat(Triticum aestivum L.)[J]. BMC Genomics, 2019, 20(1):505.
doi: 10.1186/s12864-019-5876-x pmid: 31215411 |
| [11] |
Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor[J]. Cell, 1988, 54(6):841-853.
doi: 10.1016/s0092-8674(88)91197-x pmid: 3044612 |
| [12] |
Scharf KD, Rose S, Zott W, et al. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF[J]. EMBO J, 1990, 9(13):4495-4501.
doi: 10.1002/j.1460-2075.1990.tb07900.x pmid: 2148291 |
| [13] | Sanmiya K, Koja Y, Iguchi A. The cloning of cDNAs for the heat-shock transcription factors HSFA1, HSFA2 and HSFA3 from tobacco[J]. Trop Agric Dev, 2020, 64:34-40. |
| [14] |
Wu Z, Liang JH, Wang CP, et al. Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily[J]. Plant Physiol, 2019, 181(4):1651-1667.
doi: 10.1104/pp.19.00839 URL |
| [15] |
唐锐敏, 贾小云, 朱文娇, 等. 马铃薯热激转录因子HsfA3基因的克隆及其耐热性功能分析[J]. 作物学报, 2021, 47(4):672-683.
doi: 10.3724/SP.J.1006.2021.04114 |
|
Tang RM, Jia XY, Zhu WJ, et al. Cloning of potato heat shock transcription factor StHsfA3 gene and its functional analysis in heat tolerance[J]. Acta Agron Sin, 2021, 47(4):672-683.
doi: 10.3724/SP.J.1006.2021.04114 URL |
|
| [16] |
Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature[J]. Plant Physiol, 2008, 146(2):748-761.
doi: 10.1104/pp.107.112060 pmid: 18055584 |
| [17] |
Chen H, Hwang JE, Lim CJ, et al. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response[J]. Biochem Biophys Res Commun, 2010, 401(2):238-244.
doi: 10.1016/j.bbrc.2010.09.038 URL |
| [18] |
Li ZJ, Zhang LL, Wang AX, et al. Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis[J]. PLoS One, 2013, 8(1):e54880.
doi: 10.1371/journal.pone.0054880 URL |
| [19] |
Zhu MD, Zhang M, Gao DJ, et al. Rice OsHSFA3 gene improves drought tolerance by modulating polyamine biosynthesis depending on abscisic acid and ROS levels[J]. Int J Mol Sci, 2020, 21(5):1857.
doi: 10.3390/ijms21051857 URL |
| [20] |
Mittal D, Chakrabarti S, Sarkar A, et al. Heat shock factor gene family in rice:genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses[J]. Plant Physiol Biochem, 2009, 47(9):785-795.
doi: 10.1016/j.plaphy.2009.05.003 URL |
| [21] | 吴泽. 百合热激转录因子LlHsfA3调控耐热性和耐盐性的机制解析[D]. 北京: 中国农业大学, 2018. |
| Wu Z. Analysis of thermotolerance and salt tolerance mechanism regulated by heat shock transcription factor LlHsfA3 from lily(Lili-um longiflorum)[D]. Beijing: China Agricultural University, 2018. | |
| [22] |
Song C, Chung WS, Lim CO. Overexpression of heat shock factor gene HsfA3 increases galactinol levels and oxidative stress tolerance in Arabidopsis[J]. Mol Cells, 2016, 39(6):477-483.
doi: 10.14348/molcells.2016.0027 URL |
| [23] | 庞强强, 孙晓东, 周曼, 等. 一种菜心耐热性评价方法:CN113348992A[P]. 2021-09-07. |
| Pang QQ, Sun XD, Zhou M, et al. A method for evaluating heat resistance of Chinese flowering cabbage:CN113348992A[P]. 2021-09-07. | |
| [24] | 庞强强, 周曼, 孙晓东, 等. 不同菜心品种萌发期和苗期耐热性分析及其鉴定指标筛选[J]. 西北农业学报, 2020, 29(2):295-305. |
| Pang QQ, Zhou M, Sun XD, et al. Comprehensive evaluation and indexes screening of heat tolerance at germination and seedling stages in different cultivars of Chinese flowering cabbage[J]. Acta Agric Boreali Occidentalis Sin, 2020, 29(2):295-305. | |
| [25] |
庞强强, 周曼, 孙晓东, 等. 菜心耐热性评价及酶促抗氧化系统对高温胁迫的响应[J]. 浙江农业学报, 2020, 32(1):72-79.
doi: 10.3969/j.issn.1004-1524.2020.01.09 |
| Pang QQ, Zhou M, Sun XD, et al. Evaluation of heat tolerance and response of enzymatic antioxidant system to heat stress in Brassica parachinensis L[J]. Acta Agric Zhejiangensis, 2020, 32(1):72-79. | |
| [26] | 卢宇鹏. 菜心耐热性评价及耐热基因等位变异分析[D]. 广州: 广州大学, 2020. |
| Lu YP. Evaluation of heat tolerance and analysis of allele variation of heat tolerance candidate gene in flowering Chinese cabbage[D]. Guangzhou: Guangzhou University, 2020. | |
| [27] |
Rao S, Das JR, Mathur S. Exploring the master regulator heat stress transcription factor HSFA1a-mediated transcriptional cascade of HSFs in the heat stress response of tomato[J]. J Plant Biochem Biotechnol, 2021, 30(4):878-888.
doi: 10.1007/s13562-021-00696-8 URL |
| [28] |
Meena S, Samtani H, Khurana P. Elucidating the functional role of heat stress transcription factor A6b(TaHsfA6b)in linking heat stress response and the unfolded protein response in wheat[J]. Plant Mol Biol, 2022, 108(6):621-634.
doi: 10.1007/s11103-022-01252-1 URL |
| [29] |
刘栩铭, 李敏, 段琼, 等. 蓖麻RcHSF基因家族鉴定与冷胁迫下的表达模式分析[J]. 华北农学报, 2020, 35(5):62-71.
doi: 10.7668/hbnxb.20191312 |
| Liu XM, Li M, Duan Q, et al. Identification of RcHSF gene family in castor and analysis of expression pattern under cold stress[J]. Acta Agric Boreali Sin, 2020, 35(5):62-71. | |
| [30] |
Guo M, Lu JP, Zhai YF, et al. Genome-wide analysis, expression profile of heat shock factor gene family(CaHsfs)and characterisation of CaHsfA2 in pepper(Capsicum annuum L.)[J]. BMC Plant Biol, 2015, 15:151.
doi: 10.1186/s12870-015-0512-7 URL |
| [31] |
Rehman A, Atif RM, Azhar MT, et al. Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons(Gossypium spp. )[J]. Int J Biol Macromol, 2021, 182:1507-1527.
doi: 10.1016/j.ijbiomac.2021.05.016 pmid: 33965497 |
| [32] | 张楠, 王映红, 王志敏, 等. 植物热激转录因子家族的研究进展[J]. 生物工程学报, 2021, 37(4):1155-1167. |
| Zhang N, Wang YH, Wang ZM, et al. Heat shock transcription factor family in plants:a review[J]. Chin J Biotechnol, 2021, 37(4):1155-1167. | |
| [33] | 焦淑珍, 姚文孔, 张宁波, 等. 园艺植物热激转录因子研究进展[J]. 果树学报, 2020, 37(3):419-430. |
| Jiao SZ, Yao WK, Zhang NB, et al. Research progress of heat stress transcription factors(Hsfs)in horticultural plants[J]. J Fruit Sci, 2020, 37(3):419-430. | |
| [34] | Schramm F, Larkindale J, Kiehlmann E, et al. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis[J]. The Plant Jouranl, 2008, 53(2):264-274. |
| [35] |
Yoshida T, Sakuma Y, Todaka D, et al. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system[J]. Biochem Biophys Res Commun, 2008, 368(3):515-521.
doi: 10.1016/j.bbrc.2008.01.134 URL |
| [36] | 孙天晓. 多年生黑麦草热激转录因子HSFAs和HSFCs亚家族基因的功能研究[D]. 武汉: 华中农业大学, 2021. |
| Sun TX. Function of heat shock transcription factor HSFAs and HSFCs subfamily genes in perennial ryegrass[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [37] |
Zhu XY, Huang CQ, Zhang L, et al. Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses[J]. Front Plant Sci, 2017, 8:1174.
doi: 10.3389/fpls.2017.01174 URL |
| [38] | 庞强强. 茄子HSFs和HSPs基因鉴定及其在高温下的表达模式分析[D]. 广州: 华南农业大学, 2016. |
| Pang QQ. Genome wide identification of HSFs and HSPs gene family in eggplant(Solanum melongema L.)and analysis of their expression pattern under high temperature[D]. Guangzhou: South China Agricultural University, 2016. |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [4] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [5] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [6] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [9] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [10] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [11] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [12] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [13] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [14] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| [15] | 刘媛媛, 魏传正, 谢永波, 仝宗军, 韩星, 甘炳成, 谢宝贵, 严俊杰. 金针菇II类过氧化物酶基因在子实体发育与胁迫应答过程的表达特征[J]. 生物技术通报, 2023, 39(11): 340-349. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||