








生物技术通报 ›› 2023, Vol. 39 ›› Issue (5): 217-223.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1231
姜晴春( ), 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉(
), 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉( )
)
收稿日期:2022-10-08
出版日期:2023-05-26
发布日期:2023-06-08
通讯作者:
柳忠玉,女,博士,副教授,研究方向:药用植物次生代谢与调控;E-mail: zyliu2004@126.com作者简介:姜晴春,女,硕士研究生,研究方向:植物分子生物学;E-mail: 2870140725@qq.com
基金资助:
JIANG Qing-chun( ), DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu(
), DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu( )
)
Received:2022-10-08
Published:2023-05-26
Online:2023-06-08
摘要:
对虎杖R2R3-MYB转录因子PcMYB2进行转录活性鉴定和表达特性分析,并在转基因拟南芥和转基因毛状根中进行功能研究。利用酵母单杂交试验分析PcMYB2的转录活性;实时荧光定量PCR(RT-qPCR)技术检测虎杖中PcMYB2的表达模式;DMACA法检测PcMYB2转基因拟南芥和虎杖毛状根中的原花青素含量。酵母单杂试验表明,PcMYB2具有转录激活活性;RT-qPCR结果显示,PcMYB2在虎杖根、茎、叶中均有表达,在叶中表达量最高,并且ABA和H2O2外源处理可诱导叶片中PcMYB2的表达;与野生型拟南芥相比,转基因拟南芥种皮颜色加深,原花青素含量为野生型的1.8倍;与野生型毛状根相比,转基因毛状根DMACA染色更深,原花青素含量为野生型的2.3倍。PcMYB2具有转录激活活性,且促进植物原花青素的生物合成。
姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223.
JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum[J]. Biotechnology Bulletin, 2023, 39(5): 217-223.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|
| 35Sf3 | CGGGATCCCATGGAGTCAAAGATTCA | 55 |
| 35Sr3 PcMYB2-F PcMYB2-R | AAAACTGCAGAGTCCCCCGTGTTCTCTC CGCGGATCCGTATGGGGAGAAGAAA AAAACTGCAGGAAATTTCCGAGCCATCC | 55 |
| qPcActin-F1 | GTCCCTGCCATGTATGTTGC | 58 |
| qPcActin-R1 | ACCATCACCAGAATCCAGCA | |
| qPcMYB2-F | GAGGAAGGGTGGAGAAATGA | 58 |
| qPcMYB2-R | TGCCCTTGTGCTGTATCTGA |
表1 引物序列信息
Table 1 Primer sequence information
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|
| 35Sf3 | CGGGATCCCATGGAGTCAAAGATTCA | 55 |
| 35Sr3 PcMYB2-F PcMYB2-R | AAAACTGCAGAGTCCCCCGTGTTCTCTC CGCGGATCCGTATGGGGAGAAGAAA AAAACTGCAGGAAATTTCCGAGCCATCC | 55 |
| qPcActin-F1 | GTCCCTGCCATGTATGTTGC | 58 |
| qPcActin-R1 | ACCATCACCAGAATCCAGCA | |
| qPcMYB2-F | GAGGAAGGGTGGAGAAATGA | 58 |
| qPcMYB2-R | TGCCCTTGTGCTGTATCTGA |
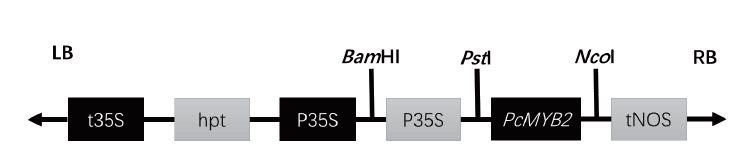
图2 pCAMBIA 1380-35S-PcMYB2载体结构示意图 LB:转移DNA的左边界;t35S:花椰菜花叶病毒CaMV 35S终止子;hpt:潮霉素抗性基因 P35S-花椰菜花叶病毒CaMV 3SS启动子; tNOS:氧化氮合酶基因终止子;RB:转移DNA的右边界
Fig. 2 Structure of pCAMBIA 1380-35S-PcMYB2 vectors LB: Left border of transfer DNA. t35S: Cauliflower mosaic virus CaMV 35S terminator. hpt: Hygromycin resistance gene P35S - Cauliflower mosaic virus CaMV 3SS promoter. tNOS: Nitric oxide synthase gene terminator. RB: Right of transfer DNA boundary
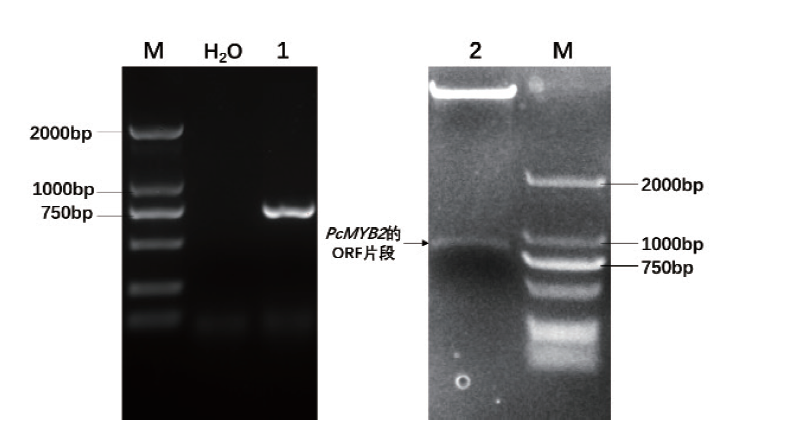
图3 重组质粒pGBKT7-PcMYB2的验证 M:Marker;1:PCR扩增获得PcMYB2基因;2:质粒pGBKT7-PcMYB2双酶切
Fig. 3 Identification of pGBKT7-PcMYB2 M: Marker. 1: PCR amplifies PcMYB2 gene. 2: Digestion pGBKT7-PcMYB2

图4 PcMYB2的转录活性验证分析 A:pGBKT7-PcMYB2转化子;B:阴性对照pGBKT7转化子
Fig. 4 Transcriptional activity verification and analysis of PcMYB2 A: Transformant pGBKT7-PcMYB2. B: Negative control transformant pGBKT7
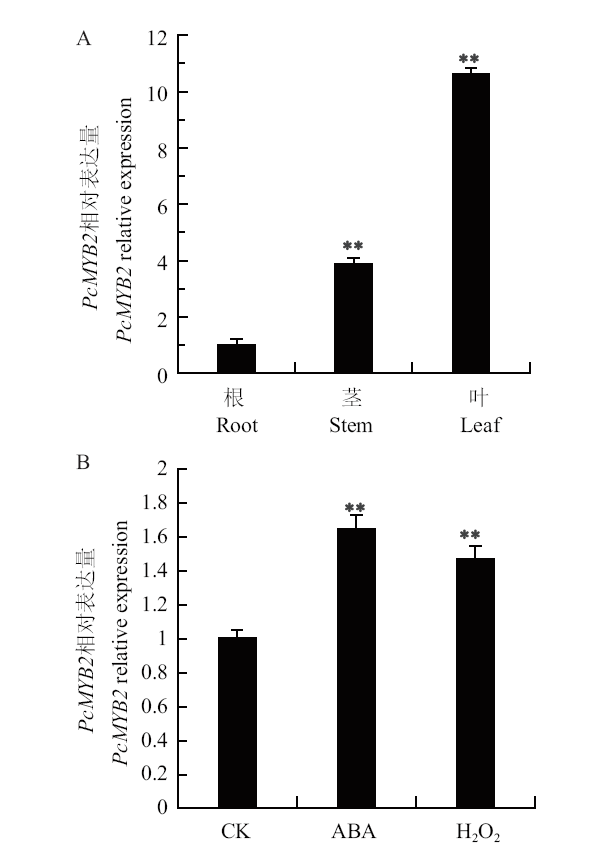
图5 PcMYB2的表达模式分析 A:不同组织PcMYB2基因表达量;B:外源信号分子处理下叶片PcMYB2的表达分析。**P< 0.01,下同
Fig. 5 Expression pattern analysis of PcMYB2 A : Expressions of PcMYB2 gene in different tissues. B : PcMYB2 expressions in leaves under exogenous signaling molecules. ** P < 0.01. The same below

图6 重组质粒pCAMBIA1380-35S-PcMYB2的验证 M:Marker;1:BamH I/ Pst I双酶切重组质粒;2: BamH I/ Nco I双酶切重组质粒
Fig. 6 Verification of recombinant plamid pCAMBIA1380-35S-PcMYB2 M: Marker. 1: Digestion recombinant plasmid by BamH I/Pst I. 2: Digestion recombinant plasmid by BamH I/ Nco I
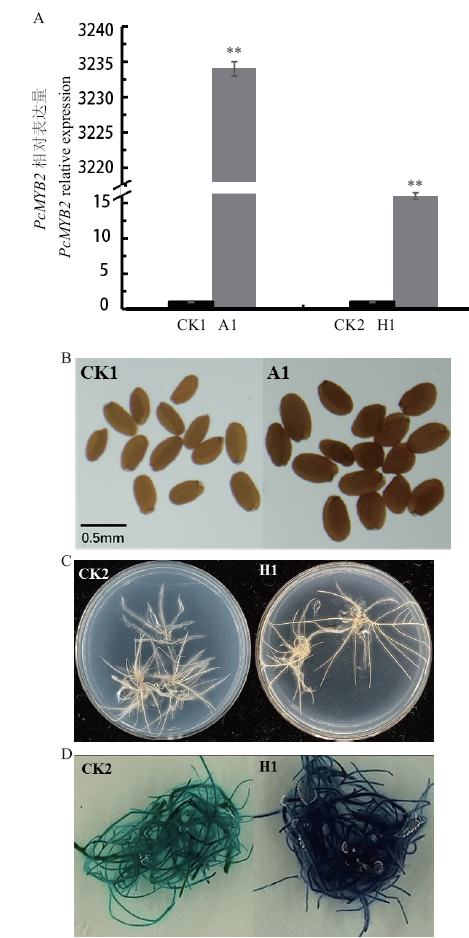
图7 转基因株系的鉴定和表现型观察 A: RT-qPCR鉴定;B:拟南芥种皮颜色观察;C: DMACA染色前的毛状根;D:DMACA染色后的毛状根。CK1:野生型拟南芥;A1:转基因拟南芥;CK2:野生型毛状根;H1:转基因毛状根
Fig. 7 Identification and phenotypic observation of trans-genic lines A: RT-qPCR identification. B: Observation on the seed coat's color of A. thaliana. C: Hairy roots before DMACA staining. D: Hairy root after DMACA staining. CK1: Wild-type A.thaliana; A1: transgenic A.thaliana; CK2: wild-type hairy roots; H1: transgenic hairy roots
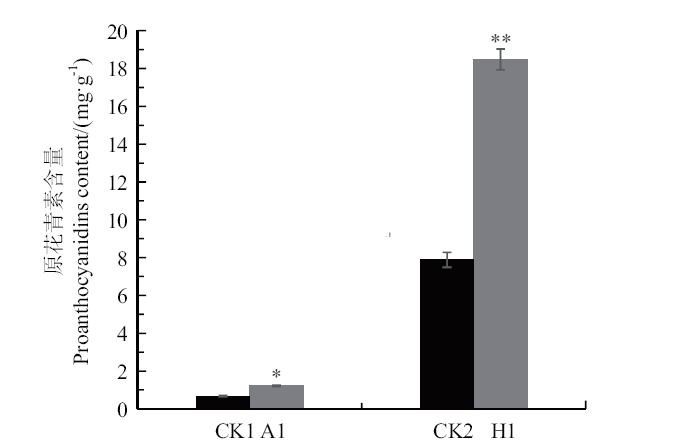
图8 原花青素含量的测定 CK1:野生型拟南芥;A1:转基因拟南芥;CK2:野生型毛状根;H1:转基因毛状根;*P < 0.05
Fig. 8 Determination of proanthocyanidins CK1: Wild-type A.thaliana; A1: transgenic A. thaliana; CK2: wild-type hairy roots; H1: transgenic hairy roots; *P < 0.05
| [1] |
Fan PH, Hostettmann K, Lou HX. Allelochemicals of the invasive neophyte Polygonum cuspidatum Sieb. & Zucc.(Polygonaceae)[J]. Chemoecology, 2010, 20(3): 223-227.
doi: 10.1007/s00049-010-0052-4 URL |
| [2] |
Li X, Liu JL, Chang QX, et al. Antioxidant and antidiabetic activity of proanthocyanidins from Fagopyrum dibotrys[J]. Molecules, 2021, 26(9): 2417.
doi: 10.3390/molecules26092417 URL |
| [3] |
Suganya M, Gnanamangai BM, Ravindran B, et al. Antitumor effect of proanthocyanidin induced apoptosis in human colorectal cancer(HT-29)cells and its molecular docking studies[J]. BMC Chem, 2019, 13(1): 21.
doi: 10.1186/s13065-019-0525-7 pmid: 31384770 |
| [4] |
Wang JY, Zhang BX, Tian ST, et al. Ectopic expression of grape hyacinth R3 MYB repressor MaMYBx affects anthocyanin and proanthocyanidin biosynthesis and epidermal cell differentiation in Arabidopsis[J]. Hortic Environ Biotechnol, 2022, 63(3): 413-423.
doi: 10.1007/s13580-021-00401-7 |
| [5] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [6] |
Zhang JK, Wang YC, Mao ZL, et al. Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple[J]. Ecotoxicol Environ Saf, 2022, 232: 113274.
doi: 10.1016/j.ecoenv.2022.113274 URL |
| [7] |
Gil-Muñoz F, Sánchez-Navarro JA, Besada C, et al. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit[J]. Sci Rep, 2020, 10(1): 3543.
doi: 10.1038/s41598-020-60635-w pmid: 32103143 |
| [8] |
Passeri V, Martens S, Carvalho E, et al. The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathway in tobacco flowers[J]. Planta, 2017, 246(2): 185-199.
doi: 10.1007/s00425-017-2667-y pmid: 28299441 |
| [9] | 李晓筱, 徐俊雄, 刘婵, 等. 虎杖MYB转录因子PcMYB2基因的克隆与原核表达[J]. 江苏农业科学, 2018, 46(23): 50-55. |
| Li XX, Xu JX, Liu S, et al. Cloning and prokaryotic expression of MYB transcription factor PcMYB2 gene from Polygonum cuspidatum[J]. Jiangsu Agric Sci, 2018, 46(23): 50-55. | |
| [10] | 房志家, 陈婷, 郝贺龙, 等. 酿酒酵母转化方法的新探索[J]. 实验室研究与探索, 2012, 31(4): 5-8, 78. |
| Fang ZJ, Chen T, Hao HL, et al. Exploration on transformation of saccharomy cescerevisiae[J]. Res Explor Lab, 2012, 31(4): 5-8, 78. | |
| [11] | 张新, 崔广艳, 陈翠娜, 等. 农杆菌介导DR1172基因转化拟南芥[J]. 基因组学与应用生物学, 2012, 31(6): 582-586. |
| Zhang X, Cui GY, Chen CN, et al. Preliminary study on Agrobacterium-mediated transformation of DR1172 gene into Arabidopsis[J]. Genom Appl Biol, 2012, 31(6): 582-586. | |
| [12] | 段梦灵, 李鲁汉, 廖辉, 等. 发根农杆菌介导的虎杖转基因体系优化[J]. 现代农业科技, 2021(4): 46-50. |
| Duan ML, Li LH, Liao H, et al. Optimization of Agrobacterium rhizogenes-mediated Polygonum cuspidatum transgenic system[J]. Mod Agric Sci Technol, 2021(4): 46-50. | |
| [13] |
Verdier J, Zhao J, Torres-Jerez I, et al. MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula[J]. Proc Natl Acad Sci USA, 2012, 109(5): 1766-1771.
doi: 10.1073/pnas.1120916109 pmid: 22307644 |
| [14] |
Wei JB, Yang JF, Jiang WB, et al. Stacking triple genes increased proanthocyanidins level in Arabidopsis thaliana[J]. PLoS One, 2020, 15(6): e0234799.
doi: 10.1371/journal.pone.0234799 URL |
| [15] |
Liu JY, Osbourn A, Ma PD. MYB transcription factors as regulators of phenylpropanoid metabolism in plants[J]. Mol Plant, 2015, 8(5): 689-708.
doi: 10.1016/j.molp.2015.03.012 pmid: 25840349 |
| [16] |
Shan TL, Rong W, Xu HJ, et al. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes[J]. Sci Rep, 2016, 6: 28777.
doi: 10.1038/srep28777 |
| [17] |
Bai QX, Duan BB, Ma JC, et al. Coexpression of PalbHLH1 and PalMYB90 genes from Populus alba enhances pathogen resistance in poplar by increasing the flavonoid content[J]. Front Plant Sci, 2020, 10: 1772.
doi: 10.3389/fpls.2019.01772 URL |
| [18] | Wang S, Wu HL, Cao XX, et al. Tartary buckwheat FtMYB30 transcription factor improves the salt/drought tolerance of transgenic Arabidopsis in an ABA-dependent manner[J]. Physiol Plant, 2022, 174(5): e13781. |
| [19] |
Wei QH, Liu YY, Lan KE, et al. Identification and analysis of MYB gene family for discovering potential regulators responding to abiotic stresses in Curcuma wenyujin[J]. Front Genet, 2022, 13: 894928.
doi: 10.3389/fgene.2022.894928 URL |
| [20] |
Wei QH, Luo QC, Wang RB, et al. A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants[J]. Front Plant Sci, 2017, 8: 1374.
doi: 10.3389/fpls.2017.01374 pmid: 28848578 |
| [21] | 王霜, 雒晓鹏, 姚英俊, 等. 苦荞R2R3-MYB转录因子调控原花青素生物合成的研究[J]. 西北植物学报, 2019, 39(11): 1911-1918. |
| Wang S, Luo XP, Yao YJ, et al. Characterization of an R2R3-MYB transcription factor involved in the synthesis of proanthocyanidins from Tartary buckwheat[J]. Acta Bot Boreali Occidentalia Sin, 2019, 39(11): 1911-1918. | |
| [22] |
Liu CG, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula[J]. Plant Physiol, 2014, 165(4): 1424-1439.
doi: 10.1104/pp.114.241877 URL |
| [23] |
Jin Z, Jiang WB, Luo YJ, et al. Analyses on flavonoids and transcriptome reveals key MYB gene for proanthocyanidins regulation in Onobrychis viciifolia[J]. Front Plant Sci, 2022, 13: 941918.
doi: 10.3389/fpls.2022.941918 URL |
| [24] |
An JP, Xu RR, Liu X, et al. Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex[J]. Plant J, 2021, 106(5): 1414-1430.
doi: 10.1111/tpj.v106.5 URL |
| [25] |
张慧文, 张玉, 马超美. 原花青素的研究进展[J]. 食品科学, 2015, 36(5): 296-304.
doi: 10.7506/spkx1002-6630-201505052 |
| Zhang HW, Zhang Y, Ma CM. Progress in procyanidins research[J]. Food Sci, 2015, 36(5): 296-304. | |
| [26] |
Luo XP, Zhao HX, Yao PF, et al. An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and proanthocyanidins from Tartary buckwheat[J]. J Plant Growth Regul, 2018, 37(1): 76-84.
doi: 10.1007/s00344-017-9709-3 URL |
| [27] |
Akagi T, Ikegami A, Yonemori K. DkMyb2 wound-induced transcription factor of persimmon(Diospyros kaki Thunb.), contributes to proanthocyanidin regulation[J]. Planta, 2010, 232(5): 1045-1059.
doi: 10.1007/s00425-010-1241-7 URL |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [4] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [5] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [6] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [7] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [8] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [9] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [10] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [11] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [12] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [13] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| [14] | 刘媛媛, 魏传正, 谢永波, 仝宗军, 韩星, 甘炳成, 谢宝贵, 严俊杰. 金针菇II类过氧化物酶基因在子实体发育与胁迫应答过程的表达特征[J]. 生物技术通报, 2023, 39(11): 340-349. |
| [15] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||