








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 26-36.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1474
收稿日期:2022-12-02
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
陈茹梅,女,研究员,研究方向:植物分子生物学与基因工程;E-mail: chenrumei@caas.cn;作者简介:李宇,女,硕士研究生,研究方向:农产品加工;E-mail: liyu397@126.com
基金资助:
LI Yu1( ), LI Su-zhen2, CHEN Ru-mei2(
), LI Su-zhen2, CHEN Ru-mei2( ), LU Hai-qiang1(
), LU Hai-qiang1( )
)
Received:2022-12-02
Published:2023-07-26
Online:2023-08-17
摘要:
铁是植物生长发育所必需的微量营养元素,生长在中性或碱性土壤中的植物普遍存在缺铁现象。而植物的正常生长发育需要保持体内铁的平衡,这种铁稳态在转录和转录后水平上都受到严格的调控。植物中铁稳态的调控网络由许多转录因子参与,其中碱性螺旋-环-螺旋(basic helix-loop helix, bHLH)家族的成员不可或缺。本文拟对植物中调控铁平衡的关键bHLH转录因子进行梳理汇总,对这些转录因子在植物生长发育中调节铁稳态的机制进行综述,以期为揭示植物铁稳态调节的研究提供理论基础。
李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36.
LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant[J]. Biotechnology Bulletin, 2023, 39(7): 26-36.
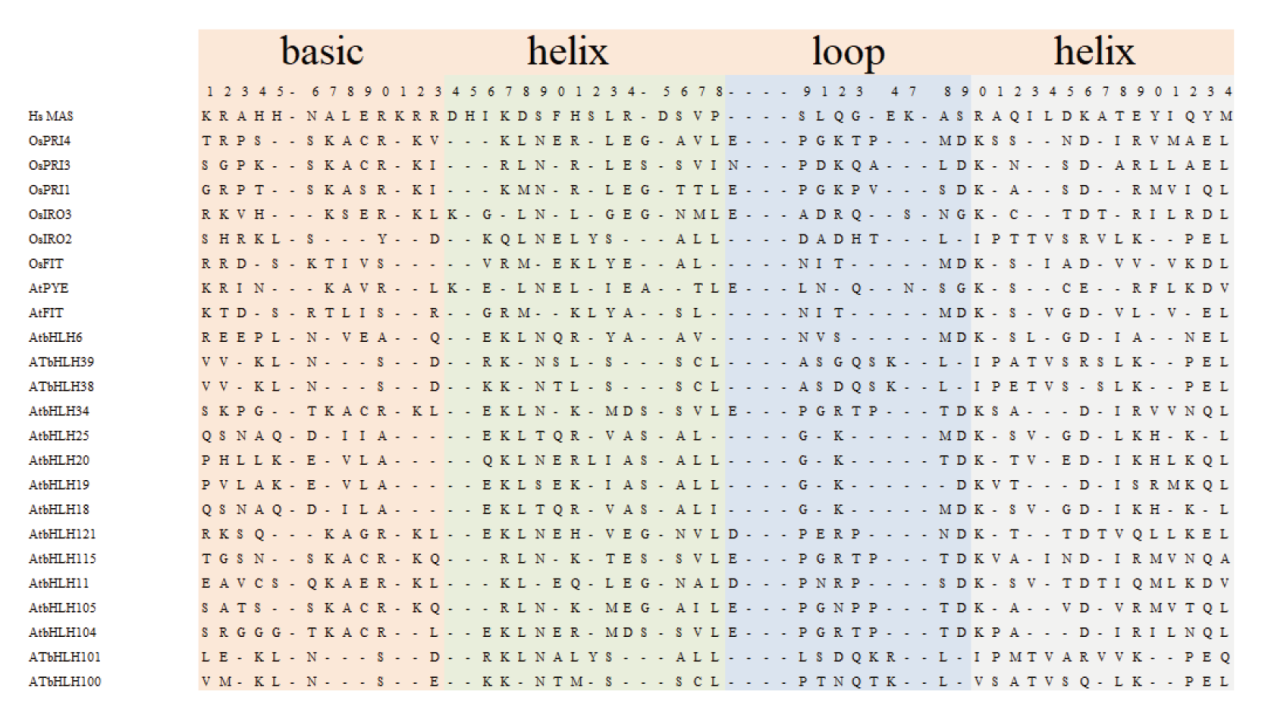
图1 铁调控网络主要蛋白bHLH结构域 图中选取了文中出现过的bHLH蛋白以及典型bHLH蛋白特性的人类蛋白MAS,阴影框里分别表示DNA结合碱性区域、两个α螺旋和一个环区
Fig. 1 Alignment of the bHLH domain of iron-regulated network main proteins The bHLH protein mentioned in the text and the typical human protein MAS with bHLH protein characteristics were selected in the figure. The shaded boxes represent the DNA binding alkaline region and two α spiral and a circular region
| [1] |
Wintz H, Fox T, Wu YY, et al. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis[J]. J Biol Chem, 2003, 278(48): 47644-47653.
doi: 10.1074/jbc.M309338200 URL |
| [2] |
Briat JF, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality[J]. Trends Plant Sci, 2015, 20(1): 33-40.
doi: 10.1016/j.tplants.2014.07.005 URL |
| [3] |
Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses[J]. Plant Physiol, 1986, 80(1): 175-180.
doi: 10.1104/pp.80.1.175 pmid: 16664577 |
| [4] |
Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants[J]. Annu Rev Plant Biol, 2012, 63: 131-152.
doi: 10.1146/annurev-arplant-042811-105522 pmid: 22404471 |
| [5] |
Gao F, Dubos C. Transcriptional integration of plant responses to iron availability[J]. J Exp Bot, 2021, 72(6): 2056-2070.
doi: 10.1093/jxb/eraa556 pmid: 33246334 |
| [6] |
Fourcroy P, Tissot N, Gaymard F, et al. Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+)transport system[J]. Mol Plant, 2016, 9(3): 485-488.
doi: S1674-2052(15)00387-1 pmid: 26415695 |
| [7] |
Robinson NJ, Procter CM, Connolly EL, et al. A ferric-chelate reductase for iron uptake from soils[J]. Nature, 1999, 397(6721): 694-697.
doi: 10.1038/17800 URL |
| [8] |
Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake[J]. Trends Plant Sci, 2015, 20(2): 124-133.
doi: 10.1016/j.tplants.2014.11.004 pmid: 25499025 |
| [9] |
Curie C, Panaviene Z, Loulergue C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III)uptake[J]. Nature, 2001, 409(6818): 346-349.
doi: 10.1038/35053080 URL |
| [10] |
Zheng LQ, Ying YH, Wang L, et al. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa[J]. BMC Plant Biol, 2010, 10: 166.
doi: 10.1186/1471-2229-10-166 |
| [11] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [12] |
Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4): 1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [13] |
Tarczewska A, Greb-Markiewicz B. The significance of the intrinsically disordered regions for the functions of the bHLH transcription factors[J]. Int J Mol Sci, 2019, 20(21): 5306.
doi: 10.3390/ijms20215306 URL |
| [14] |
Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Mol Biol Evol, 2010, 27(4): 862-874.
doi: 10.1093/molbev/msp288 pmid: 19942615 |
| [15] |
Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix(bHLH)transcription factors regulate a wide range of functions in Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 7152.
doi: 10.3390/ijms22137152 URL |
| [16] |
Filiz E, Vatansever R, Ozyigit II. Dissecting a co-expression network of basic helix-loop-helix(bHLH)genes from phosphate(Pi)-starved soybean(Glycine max)[J]. Plant Gene, 2017, 9: 19-25.
doi: 10.1016/j.plgene.2016.12.001 URL |
| [17] |
Gao F, Robe K, Gaymard F, et al. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors?[J]. Front Plant Sci, 2019, 10: 6.
doi: 10.3389/fpls.2019.00006 pmid: 30713541 |
| [18] |
Gao F, Robe K, Bettembourg M, et al. The transcription factor bHLH121 interacts with bHLH105(ILR3)and its closest homologs to regulate iron homeostasis in Arabidopsis[J]. Plant Cell, 2020, 32(2): 508-524.
doi: 10.1105/tpc.19.00541 URL |
| [19] |
Selote D, Samira R, Matthiadis A, et al. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors[J]. Plant Physiol, 2015, 167(1): 273-286.
doi: 10.1104/pp.114.250837 pmid: 25452667 |
| [20] |
Rodríguez-Celma J, Connorton JM, Kruse I, et al. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling[J]. Proc Natl Acad Sci USA, 2019, 116(35): 17584-17591.
doi: 10.1073/pnas.1907971116 pmid: 31413196 |
| [21] |
Wang N, Cui Y, Liu Y, et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana[J]. Mol Plant, 2013, 6(2): 503-513.
doi: 10.1093/mp/sss089 URL |
| [22] | Akmakjian GZ, Riaz N, Guerinot ML. Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3[J]. Proc Natl Acad Sci USA, 2021, 118(40): e2024918118. |
| [23] |
Liang G, Zhang HM, Li XL, et al. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana[J]. J Exp Bot, 2017, 68(7): 1743-1755.
doi: 10.1093/jxb/erx043 pmid: 28369511 |
| [24] |
Li XL, Zhang HM, Ai Q, et al. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana[J]. Plant Physiol, 2016, 170(4): 2478-2493.
doi: 10.1104/pp.15.01827 URL |
| [25] |
Lei RH, Li Y, Cai YR, et al. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis[J]. Mol Plant, 2020, 13(4): 634-649.
doi: 10.1016/j.molp.2020.01.006 URL |
| [26] |
Gao F, Robe K, Dubos C. Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana[J]. Plant Signal Behav, 2020, 15(10): 1795582.
doi: 10.1080/15592324.2020.1795582 URL |
| [27] |
Schwarz B, Bauer P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and-independent gene signatures[J]. J Exp Bot, 2020, 71(5): 1694-1705.
doi: 10.1093/jxb/eraa012 pmid: 31922570 |
| [28] |
Bauer P, Ling HQ, Guerinot ML. Fit, the fer-like iron deficiency induced transcription factor in Arabidopsis[J]. Plant Physiol Biochem, 2007, 45(5): 260-261.
doi: 10.1016/j.plaphy.2007.03.006 URL |
| [29] | Sivitz AB, Hermand V, Curie C, et al. Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway[J]. PLoS One, 2012, 7(9): e44843. |
| [30] | Yuan YX, Wu HL, Wang N, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis[J]. Cell Res, 2008, 18(3): 385-397. |
| [31] |
Long TA, Tsukagoshi H, Busch W, et al. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots[J]. Plant Cell, 2010, 22(7): 2219-2236.
doi: 10.1105/tpc.110.074096 URL |
| [32] |
Tanabe N, Noshi M, Mori D, et al. The basic helix-loop-helix transcription factor, bHLH11 functions in the iron-uptake system in Arabidopsis thaliana[J]. J Plant Res, 2019, 132(1): 93-105.
doi: 10.1007/s10265-018-1068-z |
| [33] |
Gratz R, Manishankar P, Ivanov R, et al. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling[J]. Dev Cell, 2019, 48(5): 726-740.e10.
doi: 10.1016/j.devcel.2019.01.006 URL |
| [34] |
Gratz R, Brumbarova T, Ivanov R, et al. Phospho-mutant activity assays provide evidence for alternative phospho-regulation pathways of the transcription factor fer-like iron deficiency-induced transcription factor[J]. New Phytol, 2020, 225(1): 250-267.
doi: 10.1111/nph.16168 pmid: 31487399 |
| [35] |
Cui Y, Chen CL, Cui M, et al. Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis[J]. Mol Plant, 2018, 11(9): 1166-1183.
doi: 10.1016/j.molp.2018.06.005 URL |
| [36] |
Matsuoka K, Furukawa J, Bidadi H, et al. Gibberellin-induced expression of Fe uptake-related genes in Arabidopsis[J]. Plant Cell Physiol, 2014, 55(1): 87-98.
doi: 10.1093/pcp/pct160 pmid: 24192296 |
| [37] |
Wild M, Davière JM, Regnault T, et al. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses[J]. Dev Cell, 2016, 37(2): 190-200.
doi: 10.1016/j.devcel.2016.03.022 URL |
| [38] |
Chao Q, Rothenberg M, Solano R, et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins[J]. Cell, 1997, 89(7): 1133-1144.
doi: 10.1016/s0092-8674(00)80300-1 pmid: 9215635 |
| [39] |
Lingam S, Mohrbacher J, Brumbarova T, et al. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis[J]. Plant Cell, 2011, 23(5): 1815-1829.
doi: 10.1105/tpc.111.084715 URL |
| [40] |
Yang Y, Ou B, Zhang JZ, et al. The Arabidopsis mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25[J]. Plant J, 2014, 77(6): 838-851.
doi: 10.1111/tpj.2014.77.issue-6 URL |
| [41] |
Zhang Y, Wu HL, Wang N, et al. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis[J]. New Phytol, 2014, 203(3): 770-783.
doi: 10.1111/nph.12860 pmid: 24889527 |
| [42] |
Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+[J]. Plant J, 2006, 45(3): 335-346.
doi: 10.1111/j.1365-313X.2005.02624.x pmid: 16412081 |
| [43] |
Zhang HM, Li Y, Pu MN, et al. Oryza sativa positive regulator of iron deficiency response 2(ospri2)and ospri3 are involved in the maintenance of fe homeostasis[J]. Plant Cell Environ, 2020, 43(1): 261-274.
doi: 10.1111/pce.v43.1 URL |
| [44] |
Kobayashi T, Nagasaka S, Senoura T, et al. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation[J]. Nat Commun, 2013, 4: 2792.
doi: 10.1038/ncomms3792 pmid: 24253678 |
| [45] |
Zhang HM, Li Y, Yao XN, et al. POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OsPRI1, facilitates iron homeostasis[J]. Plant Physiol, 2017, 175(1): 543-554.
doi: 10.1104/pp.17.00794 pmid: 28751317 |
| [46] | Wang WJ, Ye J, Ma YR, et al. OsIRO3 plays an essential role in iron deficiency responses and regulates iron homeostasis in rice[J]. Plants(Basel), 2020, 9(9): 1095. |
| [47] |
Wang WJ, Ye J, Xu H, et al. osbhlh061 links topless/topless-related repressor proteins with positive regulator of iron homeostasis 1 to maintain iron homeostasis in rice[J]. New Phytol, 2022, 234(5): 1753-1769.
doi: 10.1111/nph.18096 pmid: 35288933 |
| [48] |
Ogo Y, Itai RN, Kobayashi T, et al. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil[J]. Plant Mol Biol, 2011, 75(6): 593-605.
doi: 10.1007/s11103-011-9752-6 pmid: 21331630 |
| [49] |
Ogo Y, Itai RN, Nakanishi H, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions[J]. Plant J, 2007, 51(3): 366-377.
doi: 10.1111/j.1365-313X.2007.03149.x pmid: 17559517 |
| [50] |
Li CY, Li Y, Xu P, et al. OsIRO3 negatively regulates Fe homeostasis by repressing the expression of OsIRO2[J]. Plant J, 2022, 111(4): 966-978.
doi: 10.1111/tpj.v111.4 URL |
| [51] |
Wang SD, Li L, Ying YH, et al. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice[J]. New Phytol, 2020, 225(3): 1247-1260.
doi: 10.1111/nph.16232 pmid: 31574173 |
| [52] | Trofimov K, Ivanov R, Eutebach M, et al. Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT[J]. Plant Direct, 2019, 3(12): e00190. |
| [53] |
Brumbarova T, Bauer P. Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato[J]. Plant Physiol, 2005, 137(3): 1018-1026.
doi: 10.1104/pp.104.054270 pmid: 15695640 |
| [54] |
Bereczky Z, Wang HY, Schubert V, et al. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato[J]. J Biol Chem, 2003, 278(27): 24697-24704.
doi: 10.1074/jbc.M301365200 pmid: 12709425 |
| [55] |
Du J, Huang ZA, Wang B, et al. SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato[J]. Ann Bot, 2015, 116(1): 23-34.
doi: 10.1093/aob/mcv058 URL |
| [56] |
Zhao Q, Ren YR, Wang QJ, et al. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple[J]. Plant Biotechnol J, 2016, 14(7): 1633-1645.
doi: 10.1111/pbi.2016.14.issue-7 URL |
| [57] |
Xu HM, Wang Y, Chen F, et al. Isolation and characterization of the iron-regulated MxbHLH01 gene in Malus xiaojinensis[J]. Plant Mol Biol Rep, 2011, 29(4): 936-942.
doi: 10.1007/s11105-011-0305-6 URL |
| [58] |
Yin L, Wang Y, Yan M, et al. Molecular cloning, polyclonal antibody preparation, and characterization of a functional iron-related transcription factor IRO2 from Malus xiaojinensis[J]. Plant Physiol Biochem, 2013, 67: 63-70.
doi: 10.1016/j.plaphy.2013.02.021 URL |
| [59] | Tian Y, Pu XD, Yu HY, et al. Genome-wide characterization and analysis of bHLH transcription factors related to crocin biosynthesis in Gardenia jasminoides Ellis(Rubiaceae)[J]. Biomed Res Int, 2020, 2020: 2903861. |
| [60] |
Ling HQ, Bauer P, Bereczky Z, et al. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots[J]. Proc Natl Acad Sci USA, 2002, 99(21): 13938-13943.
doi: 10.1073/pnas.212448699 URL |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [3] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [4] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [5] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [6] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [7] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [8] | 李怡君, 吴晨晨, 李睿, 王喆, 何山文, 韦善君, 张晓霞. 水稻内生细菌新资源分离培养方案探究[J]. 生物技术通报, 2023, 39(4): 201-211. |
| [9] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [10] | 卢振万, 李雪琪, 黄金光, 周焕斌. 利用胞嘧啶碱基编辑技术创制耐草甘膦水稻[J]. 生物技术通报, 2023, 39(2): 63-69. |
| [11] | 杨茂, 林宇丰, 戴阳朔, 潘素君, 彭伟业, 严明雄, 李魏, 王冰, 戴良英. OsDIS1通过抗氧化途径负调控水稻耐旱性[J]. 生物技术通报, 2023, 39(2): 88-95. |
| [12] | 蒋铭轩, 李康, 罗亮, 刘建祥, 芦海平. 植物表达外源蛋白研究进展及展望[J]. 生物技术通报, 2023, 39(11): 110-122. |
| [13] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [14] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [15] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||