








生物技术通报 ›› 2024, Vol. 40 ›› Issue (1): 127-144.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0445
收稿日期:2023-04-12
出版日期:2024-01-26
发布日期:2024-02-06
通讯作者:
唐冬生,男,博士,教授,研究方向:基因工程与家畜分子育种;E-mail: tangdsh@163.com作者简介:杨帅朋,男,硕士研究生,研究方向:动物分子遗传与育种;E-mail: 806420672@qq.com
基金资助:
YANG Shuai-peng1( ), QU Zi-xiao1, ZHU Xiang-xing2, TANG Dong-sheng1,2(
), QU Zi-xiao1, ZHU Xiang-xing2, TANG Dong-sheng1,2( )
)
Received:2023-04-12
Published:2024-01-26
Online:2024-02-06
摘要:
碱基编辑技术起源于CRISPR/Cas系统,是目前最新的基因定点修饰技术。根据碱基编辑器的功能特点,可将碱基编辑器分为胞嘧啶碱基编辑器(cytosine base editor, CBE)、腺嘌呤碱基编辑器(adenine base editor, ABE)、糖基化酶碱基编辑器(glycosylase base editor, GBE)、腺嘌呤碱基颠换编辑器(adenine transversion base editor, AYBE)、双碱基编辑器(dual base editor, DBE)和引导编辑器(prime editor, PE)。自碱基编辑系统诞生以来,已经广泛运用于动植物的研究中,并且已经证明了它在动植物遗传改良和疾病治疗中具有巨大应用价值。猪作为一种重要的农业经济动物和优良的动物疾病模型,对其进行遗传改良则变得十分重要。碱基编辑技术因其操作便利、高效、副产物少以及性价比高等特点,被迅速应用于动植物的遗传改良,并为人类的基因治疗提供技术支持。本文着重介绍了碱基编辑技术的开发、优化、应用特点、存在的问题以及对未来的展望,并总结了其在猪中的应用。以期为相关科研工作者了解碱基编辑技术提供参考。
杨帅朋, 屈子啸, 朱向星, 唐冬生. DNA碱基编辑技术的研究进展及在猪基因修饰中的应用[J]. 生物技术通报, 2024, 40(1): 127-144.
YANG Shuai-peng, QU Zi-xiao, ZHU Xiang-xing, TANG Dong-sheng. Optimization of DNA Base Editing Technology and Its Application in Pig Genetic Modification[J]. Biotechnology Bulletin, 2024, 40(1): 127-144.
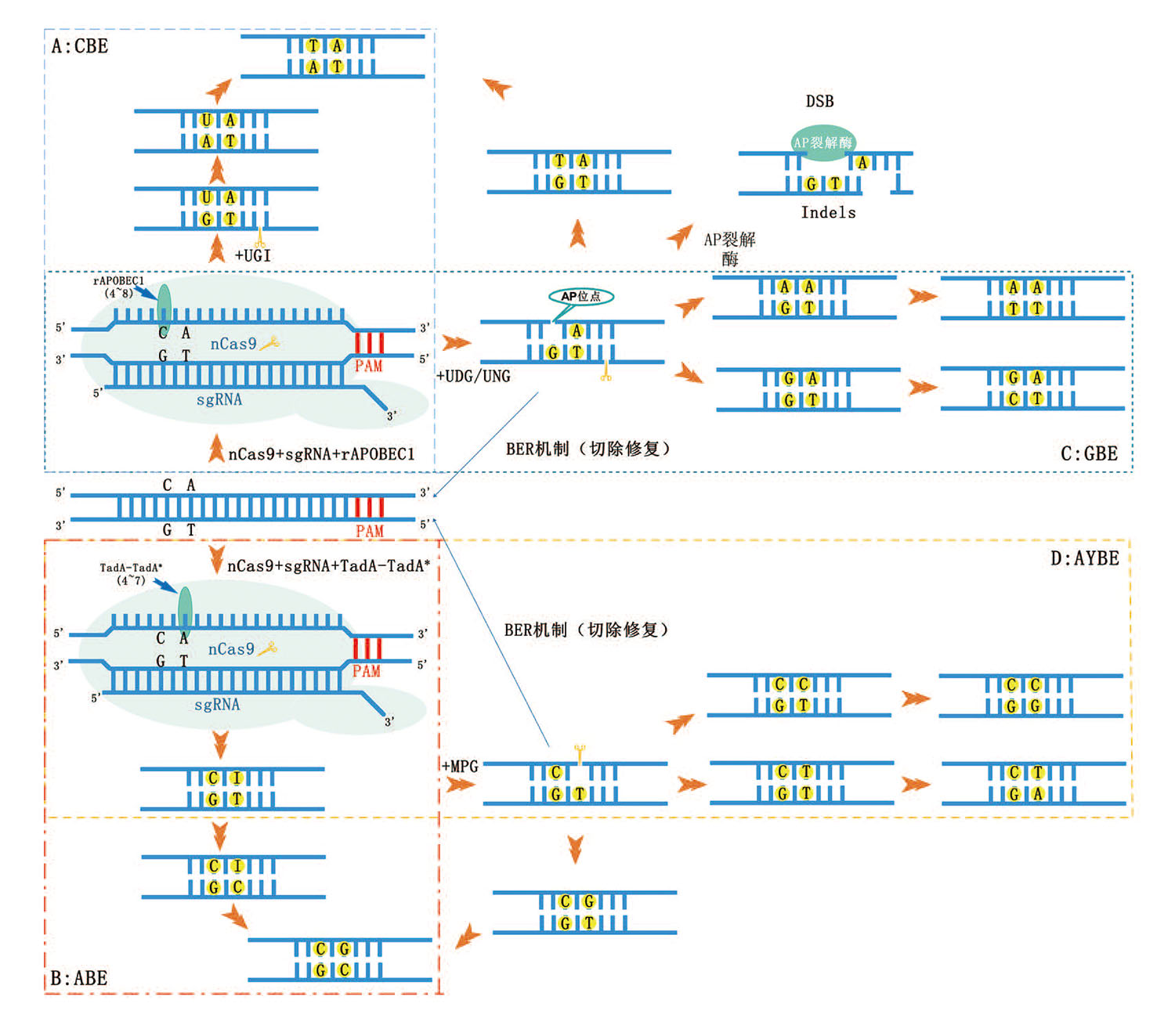
图1 碱基编辑器工作原理示意图 A:以BE3为例,碱基编辑器融合重要元件nCas9、rAPOBEC1和UGI。sgRNA和nCas9负责识别DNA特异靶点,rAPOBEC1负责将可编辑窗口的C脱氨成U,UGI抑制U变成AP状态,经过DNA复制和修复机制,最终将C G变成T A;B:以ABE7.10为例,碱基编辑器融合重要元件nCas9和TadA-TadA*。sgRNA和nCas9负责识别DNA特异靶点,TadA-TadA*负责将可编辑窗口的A脱氨成I,经过DNA复制和修复机制,最终将A T变成G C。C:以CGBE1为例,碱基编辑器融合重要元件nCas9、rAPOBEC1和UNG。sgRNA和nCas9负责识别DNA特异靶点,rAPOBEC1负责将可编辑窗口的C脱氨成U,UNG促进U位点形成AP状态,经过DNA复制和修复机制,最终将C变成A/G;D: AYBE在ABE的基础上添加MPG,A经脱氨酶形成I后,I又在MPG作用后变成AP状态,再经过DNA复制和修复机制,最终将A变成C/T。AP状态会形成Indels
Fig. 1 Mechanisms of base editor systems A: Having BE3 as an example, the base editor consists of important components nCas9, rAPOBEC1, and UGI. sgRNA and nCas9 identify specific DNA targets, then in the editable window, the rAPOBEC1 deaminates C to U, UGI inhibits U from changing to AP state, and finally changes C G to T A through DNA replication and repair mechanism. B: Having ABE7.10 as an example, the base editor consists of important components nCas9 and TadA-TadA*. sgRNA and nCas9 identify specific DNA targets, then in the editable window, the TadA-TadA* deaminates A to I, and finally changes A T into G C through DNA replication and repair mechanism. C: Having CGBE1 as an example, important components of the base editor include nCas9, then in the editable window, rAPOBEC1 deaminates C to U, and UNG. sgRNA and nCas9 identify specific DNA targets, rAPOBEC1 is responsible for deamination of C in the editable window to U, UNG promotes the formation of AP status at the U site, and finally changes C into A/G through DNA replication and repair mechanism. D: AYBE adds MPG on the basis of ABE. A becomes AP state after deamination and MPG effect, and finally becomes C/T through DNA replication and repair mechanism. After DNA replication and repair mechanism, A will finally become C/T. The AP state will form Indels
| 名称 Name | Cas蛋白 Cas protein | 工具酶 Tool enzyme | PAM序列 PAM sequence | NLS | 附加原件 Additional original | 密码子优化 Codon optimiz-ation | 编辑窗口 Editing window |
|---|---|---|---|---|---|---|---|
| BE1[ | dCas9 | rAPOBEC1 | NGG | 1× | - | - | 4-8 |
| BE2[ | dCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| BE3[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| BE3-R33A[ | nCas9 | rAPOBEC1R33A | NGG | 1× | 1×UGI | - | 5-7 |
| BE3-R33A/K34A[ | nCas9 | rAPOBEC1R33A/K34A | NGG | 1× | 1×UGI | - | 5-6 |
| Target-AID[ | nCas9 | PmCDA1 | NGG | 1× | 1×UGI | - | 2-8 |
| TAM[ | dCas9 | hAIDx | NGG | 1× | 1×UGI | - | 4-10 |
| CRISPR-X[ | dCas9 | hAIDΔ-MS2 | NGG | 1× | - | - | -50-50 |
| ZF-hAID[ | ZF | hAID | less | 1× | 1×UGI | - | - |
| TALE-hAID[ | TALE | hAID | less | 1× | 1×UGI | - | - |
| BE4[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI | - | 4-8 |
| BE4Gam[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI 1×Gam | - | 4-8 |
| eBE[ | nCas9 | rAPOBEC1 | NGG | 1× | 4×UGI | - | 4-8 |
| eA3A-BE3[ | nCas9 | hAPOBEC3A | TCR | 1× | 1×UGI | - | 7-12 |
| BE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI | - | 4-8 |
| AncBE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI | + | 4-8 |
| FNLS-BE3[ | nCas9 | rAPOBEC1 | NGG | 2× | 1×UGI | + | 3-8 |
| hyBE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI 1×Rad51DBD | - | 4-12 |
| hyA3A-BE4max[ | nCas9 | hA3A | TCR | 2× | 2×UGI 1×Rad51DBD | + | 3-15 |
| YE1-BE3[ | nCas9 | rAPOBEC1-YE1 | NGG | 1× | 1×UGI | - | 4-7 |
| YE2-BE3[ | nCas9 | rAPOBEC1-YE2 | NGG | 1× | 1×UGI | - | 5-6 |
| YEE-BE3[ | nCas9 | rAPOBEC1-YEE | NGG | 1× | 1×UGI | - | 5-6 |
| EE-BE3[ | nCas9 | rAPOBEC1-EE | NGG | 1× | 1×UGI | - | 5-5 |
| CP-CBEmax[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI | - | 4-11 |
| SaBE3[ | nSaCas9 | rAPOBEC1 | NNGRRT | 1× | 1×UGI | - | 3-12 |
| VQR-BE3[ | VQR-Cas9nCas9 | rAPOBEC1 | NGA | 1× | 1×UGI | - | 4-11 |
| EQR-BE3[ | EQR-Cas9nCas9 | rAPOBEC1 | NGAG | 1× | 1×UGI | - | 4-11 |
| VRER-BE3[ | VRER-Cas9nCas9 | rAPOBEC1 | NGCG | 1× | 1×UGI | - | 3-10 |
| SaKKH-BE3[ | nSaKKHCas9 | rAPOBEC1 | NNNRRT | 1× | 1×UGI | - | 3-12 |
| XBT3[ | nxCas9 | rAPOBEC1 | NG/GAA/GAT | 1× | 1×UGI | - | 4-8 |
| SpCas9-NG[ | nSpCas9-NG | rAPOBEC1 | NG | 1× | 1×UGI | - | 4-8 |
| dCpf1-BE[ | dCpf1(Cas12a) | rAPOBEC1 | TTTV | 3× | 4×UGI | - | 8-13 |
| enCas12a-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| RR-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| RVR-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| HF-BE3[ | HF1-nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| TBEs[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI 1×dCDI | - | 4-8 |
| SpG-CBE[ | SpG | rAPOBEC1 | NGN | 1× | 1×UGI | - | 4-8 |
| SpRY-CBE[ | SpRY | rAPOBEC1 | NRN | 1× | 1×UGI | - | 4-8 |
| ABE7.10[ | nCas9 | TadA-TadA* | NGG | 1× | - | - | 4-7 |
| ABEmax[ | nCas9 | TadA-TadA* | NGG | 2× | - | + | 4-7 |
| CP-ABEmax[ | CP-nCas9 | TadA-TadA* | NGG | 2× | - | + | 4-12 |
| ABEmaxAW[ | nCas9 | TadA-E59A-TadA*-V106W | NGG | 2× | - | + | 4-8 |
| ABEmaxQW[ | nCas9 | TadA-E59Q-TadA*-V106W | NGG | 2× | - | + | 4-8 |
| ABE7.10D53E[ | nCas9 | TadA-D53E-TadA*-D53E | NGG | 1× | - | - | 4-8 |
| ABE7.10F148A[ | nCas9 | TadA-F148A-TadA*-F148A | NGG | 1× | - | - | 5 |
| VQR-SpCas9-ABEmax[ | nVQR-SpCas9 | TadA-TadA* | NGA | 2× | - | + | 4-8 |
| VRER-SpCas9-ABEmax[ | nVRER-SpCas9 | TadA-TadA* | NGCG | 2× | - | + | 4-8 |
| VRQR-SpCas9-ABEmax[ | nVRQR-SpCas9 | TadA-TadA* | NGA | 2× | - | + | 4-8 |
| SaABEmax[ | nSaCas9 | TadA-TadA* | NNGRRT | 2× | - | + | 4-14 |
| SaKKH-ABEmax[ | nSaKKH | TadA-TadA* | NNNRRT | 2× | - | + | 4-14 |
| ScCas9-ABE7.10[ | nScCas9 | TadA-TadA* | NNGN | 1× | - | - | 4-7 |
| xABEmax[ | nxCas9 | TadA-TadA* | NG | 2× | + | 4-8 | |
| ABE-NG-S[ | SpCas9-NG | TadA* | NG | 1× | - | + | 4-7 |
| SpG-ABEmax[ | SpG | TadA-TadA* | NGN | 2× | - | - | 4-7 |
| SpRY-ABEmax[ | SpRY | TadA-TadA* | NRN | 2× | - | - | 4-7 |
| ABE8e[ | nCas9 | TadA | NGG | 1× | - | - | 3-11 |
| ABE8eV106W[ | nCas9 | TadA-V106W | NGG | 1× | - | - | 8-12 |
| enCas12a-ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| RR-ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| RVR- ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| TALE-ABE[ | TALE | TadA-F148A-TadA*-F148A | less | 1× | - | - | - |
| TALE-CBE[ | nCas9 | rAPOBEC1-YE1 | less | 1× | 1×UGI | - | - |
| GBE[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×eUNG | - | 4-8 |
| OPTI-CGBE[ | nCas9 | rAPOBEC1 | WCW | 1× | 1×UNG | + | 4-7 |
| eA3A-OPTI-CGBEs[ | nCas9 | APOBEC3A | TCN | 1× | 1×UNG | + | 4-7 |
| hA3G-OPTI-CGBEs[ | nCas9 | APOBEC3G | CCN | 1× | 1×UNG | + | 4-7 |
| hA3G-CTD-OPTI-CGBEs[ | nCas9 | APOBEC3G | CCN | 1× | 1×UNG | + | 4-7 |
| AYBEv0.1[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |
| AYBEv0.2[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |
| AYBEv0.3[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |
表1 各种碱基编辑器的特点
Table 1 Characteristics of each base editor
| 名称 Name | Cas蛋白 Cas protein | 工具酶 Tool enzyme | PAM序列 PAM sequence | NLS | 附加原件 Additional original | 密码子优化 Codon optimiz-ation | 编辑窗口 Editing window |
|---|---|---|---|---|---|---|---|
| BE1[ | dCas9 | rAPOBEC1 | NGG | 1× | - | - | 4-8 |
| BE2[ | dCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| BE3[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| BE3-R33A[ | nCas9 | rAPOBEC1R33A | NGG | 1× | 1×UGI | - | 5-7 |
| BE3-R33A/K34A[ | nCas9 | rAPOBEC1R33A/K34A | NGG | 1× | 1×UGI | - | 5-6 |
| Target-AID[ | nCas9 | PmCDA1 | NGG | 1× | 1×UGI | - | 2-8 |
| TAM[ | dCas9 | hAIDx | NGG | 1× | 1×UGI | - | 4-10 |
| CRISPR-X[ | dCas9 | hAIDΔ-MS2 | NGG | 1× | - | - | -50-50 |
| ZF-hAID[ | ZF | hAID | less | 1× | 1×UGI | - | - |
| TALE-hAID[ | TALE | hAID | less | 1× | 1×UGI | - | - |
| BE4[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI | - | 4-8 |
| BE4Gam[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI 1×Gam | - | 4-8 |
| eBE[ | nCas9 | rAPOBEC1 | NGG | 1× | 4×UGI | - | 4-8 |
| eA3A-BE3[ | nCas9 | hAPOBEC3A | TCR | 1× | 1×UGI | - | 7-12 |
| BE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI | - | 4-8 |
| AncBE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI | + | 4-8 |
| FNLS-BE3[ | nCas9 | rAPOBEC1 | NGG | 2× | 1×UGI | + | 3-8 |
| hyBE4max[ | nCas9 | rAPOBEC1 | NGG | 2× | 2×UGI 1×Rad51DBD | - | 4-12 |
| hyA3A-BE4max[ | nCas9 | hA3A | TCR | 2× | 2×UGI 1×Rad51DBD | + | 3-15 |
| YE1-BE3[ | nCas9 | rAPOBEC1-YE1 | NGG | 1× | 1×UGI | - | 4-7 |
| YE2-BE3[ | nCas9 | rAPOBEC1-YE2 | NGG | 1× | 1×UGI | - | 5-6 |
| YEE-BE3[ | nCas9 | rAPOBEC1-YEE | NGG | 1× | 1×UGI | - | 5-6 |
| EE-BE3[ | nCas9 | rAPOBEC1-EE | NGG | 1× | 1×UGI | - | 5-5 |
| CP-CBEmax[ | nCas9 | rAPOBEC1 | NGG | 1× | 2×UGI | - | 4-11 |
| SaBE3[ | nSaCas9 | rAPOBEC1 | NNGRRT | 1× | 1×UGI | - | 3-12 |
| VQR-BE3[ | VQR-Cas9nCas9 | rAPOBEC1 | NGA | 1× | 1×UGI | - | 4-11 |
| EQR-BE3[ | EQR-Cas9nCas9 | rAPOBEC1 | NGAG | 1× | 1×UGI | - | 4-11 |
| VRER-BE3[ | VRER-Cas9nCas9 | rAPOBEC1 | NGCG | 1× | 1×UGI | - | 3-10 |
| SaKKH-BE3[ | nSaKKHCas9 | rAPOBEC1 | NNNRRT | 1× | 1×UGI | - | 3-12 |
| XBT3[ | nxCas9 | rAPOBEC1 | NG/GAA/GAT | 1× | 1×UGI | - | 4-8 |
| SpCas9-NG[ | nSpCas9-NG | rAPOBEC1 | NG | 1× | 1×UGI | - | 4-8 |
| dCpf1-BE[ | dCpf1(Cas12a) | rAPOBEC1 | TTTV | 3× | 4×UGI | - | 8-13 |
| enCas12a-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| RR-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| RVR-A3A-Y130F[ | LbCas12a | hA3A | NTTN/TYCN/TRTV | 1× | 1×UGI | + | 7-12 |
| HF-BE3[ | HF1-nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI | - | 4-8 |
| TBEs[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×UGI 1×dCDI | - | 4-8 |
| SpG-CBE[ | SpG | rAPOBEC1 | NGN | 1× | 1×UGI | - | 4-8 |
| SpRY-CBE[ | SpRY | rAPOBEC1 | NRN | 1× | 1×UGI | - | 4-8 |
| ABE7.10[ | nCas9 | TadA-TadA* | NGG | 1× | - | - | 4-7 |
| ABEmax[ | nCas9 | TadA-TadA* | NGG | 2× | - | + | 4-7 |
| CP-ABEmax[ | CP-nCas9 | TadA-TadA* | NGG | 2× | - | + | 4-12 |
| ABEmaxAW[ | nCas9 | TadA-E59A-TadA*-V106W | NGG | 2× | - | + | 4-8 |
| ABEmaxQW[ | nCas9 | TadA-E59Q-TadA*-V106W | NGG | 2× | - | + | 4-8 |
| ABE7.10D53E[ | nCas9 | TadA-D53E-TadA*-D53E | NGG | 1× | - | - | 4-8 |
| ABE7.10F148A[ | nCas9 | TadA-F148A-TadA*-F148A | NGG | 1× | - | - | 5 |
| VQR-SpCas9-ABEmax[ | nVQR-SpCas9 | TadA-TadA* | NGA | 2× | - | + | 4-8 |
| VRER-SpCas9-ABEmax[ | nVRER-SpCas9 | TadA-TadA* | NGCG | 2× | - | + | 4-8 |
| VRQR-SpCas9-ABEmax[ | nVRQR-SpCas9 | TadA-TadA* | NGA | 2× | - | + | 4-8 |
| SaABEmax[ | nSaCas9 | TadA-TadA* | NNGRRT | 2× | - | + | 4-14 |
| SaKKH-ABEmax[ | nSaKKH | TadA-TadA* | NNNRRT | 2× | - | + | 4-14 |
| ScCas9-ABE7.10[ | nScCas9 | TadA-TadA* | NNGN | 1× | - | - | 4-7 |
| xABEmax[ | nxCas9 | TadA-TadA* | NG | 2× | + | 4-8 | |
| ABE-NG-S[ | SpCas9-NG | TadA* | NG | 1× | - | + | 4-7 |
| SpG-ABEmax[ | SpG | TadA-TadA* | NGN | 2× | - | - | 4-7 |
| SpRY-ABEmax[ | SpRY | TadA-TadA* | NRN | 2× | - | - | 4-7 |
| ABE8e[ | nCas9 | TadA | NGG | 1× | - | - | 3-11 |
| ABE8eV106W[ | nCas9 | TadA-V106W | NGG | 1× | - | - | 8-12 |
| enCas12a-ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| RR-ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| RVR- ABE8eV106W[ | LbCas12a | TadA-V106W | TTTV | 1× | - | + | 8-12 |
| TALE-ABE[ | TALE | TadA-F148A-TadA*-F148A | less | 1× | - | - | - |
| TALE-CBE[ | nCas9 | rAPOBEC1-YE1 | less | 1× | 1×UGI | - | - |
| GBE[ | nCas9 | rAPOBEC1 | NGG | 1× | 1×eUNG | - | 4-8 |
| OPTI-CGBE[ | nCas9 | rAPOBEC1 | WCW | 1× | 1×UNG | + | 4-7 |
| eA3A-OPTI-CGBEs[ | nCas9 | APOBEC3A | TCN | 1× | 1×UNG | + | 4-7 |
| hA3G-OPTI-CGBEs[ | nCas9 | APOBEC3G | CCN | 1× | 1×UNG | + | 4-7 |
| hA3G-CTD-OPTI-CGBEs[ | nCas9 | APOBEC3G | CCN | 1× | 1×UNG | + | 4-7 |
| AYBEv0.1[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |
| AYBEv0.2[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |
| AYBEv0.3[ | nCas9 | TadA | NGG | 1× | 1×MPG | + | 3-11 |

图2 引导编辑器工作原理示意图 以PE2为例,碱基编辑器融合重要元件PE2蛋白(nCas9(H840A)+RTase)和pegRNA(sgRNA+RTT+PBS)。sgRNA和nCas9(H840A)负责识别特异DNA序列和切割单链,RTT和PBS负责提供逆转录结合位点和逆转录模板,经过DNA复制和修复机制,最终对目标靶点进行定向改造
Fig. 2 Mechanisms of prime editors Having PE2 as an example, the base editor is composed of important components PE2 protein(nCas9(H840A)+RTase)and pegRNA(sgRNA+RTT+PBS). sgRNA and nCas9(H840A)recognize specific DNA sequences and cut single strands. RTT and PBS provide reverse transcription binding sites and reverse transcription templates. After DNA replication and repair mechanisms, target targets are finally targeted for modification
| [1] |
Kim JS. Genome editing comes of age[J]. Nat Protoc, 2016, 11(9): 1573-1578.
doi: 10.1038/nprot.2016.104 |
| [2] |
Danna K, Nathans D. Specific cleavage of Simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae[J]. Proc Natl Acad Sci USA, 1971, 68(12): 2913-2917.
pmid: 4332003 |
| [3] |
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain[J]. Proc Natl Acad Sci USA, 1996, 93(3): 1156-1160.
doi: 10.1073/pnas.93.3.1156 pmid: 8577732 |
| [4] |
Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases[J]. Genetics, 2010, 186(2): 757-761.
doi: 10.1534/genetics.110.120717 pmid: 20660643 |
| [5] | Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria[J]. Proc Natl Acad Sci USA, 2012, 109(39): E2579-E2586. |
| [6] |
Bibikova M, Golic M, Golic KG, et al. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases[J]. Genetics, 2002, 161(3): 1169-1175.
doi: 10.1093/genetics/161.3.1169 pmid: 12136019 |
| [7] |
Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors[J]. Science, 2009, 326(5959): 1509-1512.
doi: 10.1126/science.1178811 pmid: 19933107 |
| [8] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096.
doi: 10.1126/science.1258096 URL |
| [9] |
Gao CX. Genome engineering for crop improvement and future agriculture[J]. Cell, 2021, 184(6): 1621-1635.
doi: 10.1016/j.cell.2021.01.005 pmid: 33581057 |
| [10] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278.
doi: S0092-8674(14)00604-7 pmid: 24906146 |
| [11] |
Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells[J]. Nat Rev Genet, 2018, 19(12): 770-788.
doi: 10.1038/s41576-018-0059-1 |
| [12] |
Hess GT, Tycko J, Yao D, et al. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes[J]. Mol Cell, 2017, 68(1): 26-43.
doi: S1097-2765(17)30707-4 pmid: 28985508 |
| [13] |
Karanam K, Kafri R, Loewer A, et al. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase[J]. Mol Cell, 2012, 47(2): 320-329.
doi: 10.1016/j.molcel.2012.05.052 pmid: 22841003 |
| [14] |
Zhang Y, Massel K, Godwin ID, et al. Applications and potential of genome editing in crop improvement[J]. Genome Biol, 2018, 19(1): 210.
doi: 10.1186/s13059-018-1586-y pmid: 30501614 |
| [15] |
Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants[J]. Nucleic Acids Res, 2016, 44(D1): D862-D868.
doi: 10.1093/nar/gkv1222 URL |
| [16] |
Zhao KY, Tung CW, Eizenga GC, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa[J]. Nat Commun, 2011, 2: 467.
doi: 10.1038/ncomms1467 |
| [17] |
Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 |
| [18] |
Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 URL |
| [19] |
Koblan LW, Doman JL, Wilson C, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction[J]. Nat Biotechnol, 2018, 36(9): 843-846.
doi: 10.1038/nbt.4172 pmid: 29813047 |
| [20] |
Zhao DD, Li J, Li SW, et al. Glycosylase base editors enable C-to-A and C-to-G base changes[J]. Nat Biotechnol, 2021, 39(1): 35-40.
doi: 10.1038/s41587-020-0592-2 |
| [21] |
Tong HW, Wang XC, Liu YH, et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase[J]. Nat Biotechnol, 2023, 41(8): 1080-1084.
doi: 10.1038/s41587-022-01595-6 pmid: 36624150 |
| [22] |
Li C, Zhang R, Meng XB, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors[J]. Nat Biotechnol, 2020, 38(7): 875-882.
doi: 10.1038/s41587-019-0393-7 pmid: 31932727 |
| [23] |
Zhang XH, Zhu BY, Chen L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells[J]. Nat Biotechnol, 2020, 38(7): 856-860.
doi: 10.1038/s41587-020-0527-y pmid: 32483363 |
| [24] |
Grünewald J, Zhou RH, Lareau CA, et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing[J]. Nat Biotechnol, 2020, 38(7): 861-864.
doi: 10.1038/s41587-020-0535-y pmid: 32483364 |
| [25] |
Sakata RC, Ishiguro S, Mori H, et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations[J]. Nat Biotechnol, 2020, 38(7): 865-869.
doi: 10.1038/s41587-020-0509-0 pmid: 32483365 |
| [26] |
Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157.
doi: 10.1038/s41586-019-1711-4 |
| [27] | Wei Y, Zhang XH, Li DL. The new favorite of gene editing technology-single base editors[J]. Yi Chuan, 2017, 39(12): 1115-1121. |
| [28] |
Grünewald J, Zhou RH, Garcia SP, et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors[J]. Nature, 2019, 569(7756): 433-437.
doi: 10.1038/s41586-019-1161-z |
| [29] |
Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): aaf8729.
doi: 10.1126/science.aaf8729 URL |
| [30] |
Ma YQ, Zhang JY, Yin WJ, et al. Targeted AID-mediated mutagenesis(TAM)enables efficient genomic diversification in mammalian cells[J]. Nat Methods, 2016, 13(12): 1029-1035.
doi: 10.1126/science.13.339.1029.b URL |
| [31] |
Hess GT, Frésard L, Han K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells[J]. Nat Methods, 2016, 13(12): 1036-1042.
doi: 10.1038/nmeth.4038 pmid: 27798611 |
| [32] |
Yang LH, Briggs AW, Chew WL, et al. Engineering and optimising deaminase fusions for genome editing[J]. Nat Commun, 2016, 7: 13330.
doi: 10.1038/ncomms13330 pmid: 27804970 |
| [33] |
Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C: G-to-T: a base editors with higher efficiency and product purity[J]. Sci Adv, 2017, 3(8): eaao4774.
doi: 10.1126/sciadv.aao4774 URL |
| [34] |
Wang LJ, Xue W, Yan L, et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor[J]. Cell Res, 2017, 27(10): 1289-1292.
doi: 10.1038/cr.2017.111 pmid: 28849781 |
| [35] |
Gehrke JM, Cervantes O, Clement MK, et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities[J]. Nat Biotechnol, 2018, 36(10): 977-982.
doi: 10.1038/nbt.4199 pmid: 30059493 |
| [36] |
Zafra MP, Schatoff EM, Katti A, et al. Optimized base editors enable efficient editing in cells, organoids and mice[J]. Nat Biotechnol, 2018, 36(9): 888-893.
doi: 10.1038/nbt.4194 pmid: 29969439 |
| [37] |
Zhang XH, Chen L, Zhu BY, et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain[J]. Nat Cell Biol, 2020, 22(6): 740-750.
doi: 10.1038/s41556-020-0518-8 pmid: 32393889 |
| [38] |
Kim YB, Komor AC, Levy JM, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions[J]. Nat Biotechnol, 2017, 35(4): 371-376.
doi: 10.1038/nbt.3803 pmid: 28191901 |
| [39] |
Huang TP, Zhao KT, Miller SM, et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors[J]. Nat Biotechnol, 2019, 37(6): 626-631.
doi: 10.1038/s41587-019-0134-y pmid: 31110355 |
| [40] |
Nishimasu H, Shi X, Ishiguro S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space[J]. Science, 2018, 361(6408): 1259-1262.
doi: 10.1126/science.aas9129 pmid: 30166441 |
| [41] |
Li XS, Wang Y, Liu YJ, et al. Base editing with a Cpf1-cytidine deaminase fusion[J]. Nat Biotechnol, 2018, 36(4): 324-327.
doi: 10.1038/nbt.4102 pmid: 29553573 |
| [42] |
Chen FB, Lian M, Ma BX, et al. Multiplexed base editing through Cas12a variant-mediated cytosine and adenine base editors[J]. Commun Biol, 2022, 5(1): 1163.
doi: 10.1038/s42003-022-04152-8 pmid: 36323848 |
| [43] |
Rees HA, Komor AC, Yeh WH, et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery[J]. Nat Commun, 2017, 8: 15790.
doi: 10.1038/ncomms15790 pmid: 28585549 |
| [44] |
Wang LJ, Xue W, Zhang HX, et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations[J]. Nat Cell Biol, 2021, 23(5): 552-563.
doi: 10.1038/s41556-021-00671-4 pmid: 33972728 |
| [45] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [46] |
Rees HA, Wilson C, Doman JL, et al. Analysis and minimization of cellular RNA editing by DNA adenine base editors[J]. Sci Adv, 2019, 5(5): eaax5717.
doi: 10.1126/sciadv.aax5717 URL |
| [47] |
Zhou CY, Sun YD, Yan R, et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis[J]. Nature, 2019, 571(7764): 275-278.
doi: 10.1038/s41586-019-1314-0 |
| [48] |
Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog[J]. Sci Adv, 2018, 4(10): eaau0766.
doi: 10.1126/sciadv.aau0766 URL |
| [49] |
Hua K, Tao XP, Han PJ, et al. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences[J]. Mol Plant, 2019, 12(7): 1003-1014.
doi: S1674-2052(19)30122-4 pmid: 30928636 |
| [50] |
Richter MF, Zhao KT, Eton E, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity[J]. Nat Biotechnol, 2020, 38(7): 883-891.
doi: 10.1038/s41587-020-0453-z pmid: 32433547 |
| [51] |
Liu Y, Zhou JZ, Lan T, et al. Elimination of Cas9-dependent off-targeting of adenine base editor by using TALE to separately guide deaminase to target sites[J]. Cell Discov, 2022, 8(1): 28.
doi: 10.1038/s41421-022-00384-4 pmid: 35322006 |
| [52] |
Zhou JZ, Liu Y, Wei YH, et al. Eliminating predictable DNA off-target effects of cytosine base editor by using dual guiders including sgRNA and TALE[J]. Mol Ther, 2022, 30(7): 2443-2451.
doi: 10.1016/j.ymthe.2022.04.010 pmid: 35443934 |
| [53] |
Kurt IC, Zhou RH, Iyer S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells[J]. Nat Biotechnol, 2021, 39(1): 41-46.
doi: 10.1038/s41587-020-0609-x |
| [54] |
Yuan TL, Yan NN, Fei TY, et al. Optimization of C-to-G base editors with sequence context preference predictable by machine learning methods[J]. Nat Commun, 2021, 12(1): 4902.
doi: 10.1038/s41467-021-25217-y pmid: 34385461 |
| [55] |
Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements[J]. Nat Biotechnol, 2018, 36(8): 765-771.
doi: 10.1038/nbt.4192 pmid: 30010673 |
| [56] |
Cullot G, Boutin J, Toutain J, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations[J]. Nat Commun, 2019, 10(1): 1136.
doi: 10.1038/s41467-019-09006-2 pmid: 30850590 |
| [57] |
Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells[J]. Nat Med, 2018, 24(7): 939-946.
doi: 10.1038/s41591-018-0050-6 pmid: 29892062 |
| [58] |
Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63.
doi: 10.1038/nature26155 URL |
| [59] |
Jin S, Zong Y, Gao Q, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice[J]. Science, 2019, 364(6437): 292-295.
doi: 10.1126/science.aaw7166 pmid: 30819931 |
| [60] |
Zuo EW, Sun YD, Wei W, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos[J]. Science, 2019, 364(6437): 289-292.
doi: 10.1126/science.aav9973 pmid: 30819928 |
| [61] |
Kim D, Kim DE, Lee G, et al. Genome-wide target specificity of CRISPR RNA-guided adenine base editors[J]. Nat Biotechnol, 2019, 37(4): 430-435.
doi: 10.1038/s41587-019-0050-1 pmid: 30833658 |
| [62] |
Kim D, Lim K, Kim ST, et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases[J]. Nat Biotechnol, 2017, 35(5): 475-480.
doi: 10.1038/nbt.3852 pmid: 28398345 |
| [63] |
Lee JK, Jeong E, Lee J, et al. Directed evolution of CRISPR-Cas9 to increase its specificity[J]. Nat Commun, 2018, 9(1): 3048.
doi: 10.1038/s41467-018-05477-x pmid: 30082838 |
| [64] |
Zong Y, Song QN, Li C, et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A[J]. Nat Biotechnol, 2018: 36(9): 950-953.
doi: 10.1038/nbt.4261 URL |
| [65] |
Ren B, Yan F, Kuang YJ, et al. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant[J]. Mol Plant, 2018, 11(4): 623-626.
doi: S1674-2052(18)30025-X pmid: 29382569 |
| [66] |
Wang X, Li JN, Wang Y, et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion[J]. Nat Biotechnol, 2018, 36(10): 946-949.
doi: 10.1038/nbt.4198 pmid: 30125268 |
| [67] |
Yang B, Yang L, Chen J. Development and application of base editors[J]. CRISPR J, 2019, 2(2): 91-104.
doi: 10.1089/crispr.2019.0001 pmid: 30998092 |
| [68] |
Hua K, Tao XP, Zhu JK. Expanding the base editing scope in rice by using Cas9 variants[J]. Plant Biotechnol J, 2019, 17(2): 499-504.
doi: 10.1111/pbi.12993 pmid: 30051586 |
| [69] |
Lapinaite A, Knott GJ, Palumbo CM, et al. DNA capture by a CRISPR-Cas9-guided adenine base editor[J]. Science, 2020, 369(6503): 566-571.
doi: 10.1126/science.abb1390 pmid: 32732424 |
| [70] |
Koblan LW, Arbab M, Shen MW, et al. Efficient C·G-to-G·C base editors developed using CRISPRi screens, target-library analysis, and machine learning[J]. Nat Biotechnol, 2021, 39(11): 1414-1425.
doi: 10.1038/s41587-021-00938-z pmid: 34183861 |
| [71] |
Martín-Alonso S, Frutos-Beltrán E, Menéndez-Arias L. Reverse transcriptase: from transcriptomics to genome editing[J]. Trends Biotechnol, 2021, 39(2): 194-210.
doi: 10.1016/j.tibtech.2020.06.008 pmid: 32653101 |
| [72] |
Liu Y, Yang G, Huang SH, et al. Enhancing prime editing by Csy4-mediated processing of pegRNA[J]. Cell Res, 2021, 31(10): 1134-1136.
doi: 10.1038/s41422-021-00520-x pmid: 34103663 |
| [73] |
Nelson JW, Randolph PB, Shen SP, et al. Engineered pegRNAs improve prime editing efficiency[J]. Nat Biotechnol, 2022, 40(3): 402-410.
doi: 10.1038/s41587-021-01039-7 |
| [74] |
Zhang GQ, Liu Y, Huang SS, et al. Enhancement of prime editing via xrRNA motif-joined pegRNA[J]. Nat Commun, 2022, 13(1): 1856.
doi: 10.1038/s41467-022-29507-x pmid: 35387980 |
| [75] |
Li XY, Wang X, Sun WJ, et al. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes[J]. J Mol Cell Biol, 2022, 14(4): mjac022.
doi: 10.1093/jmcb/mjac022 URL |
| [76] |
Li XS, Zhou LN, Gao BQ, et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure[J]. Nat Commun, 2022, 13(1): 1669.
doi: 10.1038/s41467-022-29339-9 pmid: 35351879 |
| [77] |
Lin QP, Jin S, Zong Y, et al. High-efficiency prime editing with optimized, paired pegRNAs in plants[J]. Nat Biotechnol, 2021, 39(8): 923-927.
doi: 10.1038/s41587-021-00868-w pmid: 33767395 |
| [78] |
Zhuang Y, Liu JL, Wu H, et al. Increasing the efficiency and precision of prime editing with guide RNA pairs[J]. Nat Chem Biol, 2022, 18(1): 29-37.
doi: 10.1038/s41589-021-00889-1 |
| [79] |
Song M, Lim JM, Min S, et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain[J]. Nat Commun, 2021, 12(1): 5617.
doi: 10.1038/s41467-021-25928-2 pmid: 34556671 |
| [80] |
Park SJ, Jeong TY, Shin SK, et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor[J]. Genome Biol, 2021, 22(1): 170.
doi: 10.1186/s13059-021-02389-w |
| [81] |
Chen PJ, Hussmann JA, Yan J, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes[J]. Cell, 2021, 184(22): 5635-5652.e29.
doi: 10.1016/j.cell.2021.09.018 pmid: 34653350 |
| [82] |
Xu W, Yang YX, Yang BY, et al. A design optimized prime editor with expanded scope and capability in plants[J]. Nat Plants, 2022, 8(1): 45-52.
doi: 10.1038/s41477-021-01043-4 |
| [83] |
Zong Y, Liu YJ, Xue CX, et al. An engineered prime editor with enhanced editing efficiency in plants[J]. Nat Biotechnol, 2022, 40(9): 1394-1402.
doi: 10.1038/s41587-022-01254-w pmid: 35332341 |
| [84] |
Ferreira da Silva J, Oliveira GP, Arasa-Verge EA, et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair[J]. Nat Commun, 2022, 13(1): 760.
doi: 10.1038/s41467-022-28442-1 pmid: 35140211 |
| [85] |
Liu PP, Liang SQ, Zheng CW, et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice[J]. Nat Commun, 2021, 12(1): 2121.
doi: 10.1038/s41467-021-22295-w pmid: 33837189 |
| [86] |
Lu YM, Tian YF, Shen RD, et al. Precise genome modification in tomato using an improved prime editing system[J]. Plant Biotechnol J, 2021, 19(3): 415-417.
doi: 10.1111/pbi.13497 pmid: 33091225 |
| [87] |
Kweon J, Yoon JK, Jang AH, et al. Engineered prime editors with PAM flexibility[J]. Mol Ther, 2021, 29(6): 2001-2007.
doi: 10.1016/j.ymthe.2021.02.022 pmid: 33636398 |
| [88] |
Oh Y, Lee WJ, Hur JK, et al. Expansion of the prime editing modality with Cas9 from Francisella novicida[J]. Genome Biol, 2022, 23(1): 92.
doi: 10.1186/s13059-022-02644-8 |
| [89] |
Anzalone AV, Gao XD, Podracky CJ, et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing[J]. Nat Biotechnol, 2022, 40(5): 731-740.
doi: 10.1038/s41587-021-01133-w |
| [90] |
Choi J, Chen W, Suiter CC, et al. Precise genomic deletions using paired prime editing[J]. Nat Biotechnol, 2022, 40(2): 218-226.
doi: 10.1038/s41587-021-01025-z |
| [91] |
Wang JL, He Z, Wang GQ, et al. Efficient targeted insertion of large DNA fragments without DNA donors[J]. Nat Methods, 2022, 19(3): 331-340.
doi: 10.1038/s41592-022-01399-1 pmid: 35228726 |
| [92] | Tao R, Wang YH, Hu Y, et al. WT-PE: prime editing with nuclease wild-type Cas9 enables versatile large-scale genome editing[J]. Signal Transduct Target Ther, 2022, 7(1): 108. |
| [93] |
Wang Y, Bi DF, Qin GS, et al. Cytosine base editor(hA3A-BE3-NG)-mediated multiple gene editing for pyramid breeding in pigs[J]. Front Genet, 2020, 11: 592623.
doi: 10.3389/fgene.2020.592623 URL |
| [94] |
Pan JS, Lin ZS, Wen JC, et al. Application of the modified cytosine base-editing in the cultured cells of Bama minipig[J]. Biotechnol Lett, 2021, 43(9): 1699-1714.
doi: 10.1007/s10529-021-03159-1 |
| [95] | 赵为民, 王慧利, 曹少先, 等. 猪CD163基因的单碱基编辑研究[J]. 畜牧兽医学报, 2022, 53(4): 1041-1050. |
| Zhao WM, Wang HL, Cao SX, et al. The study of base editing of porcine CD163 gene[J]. Acta Vet Zootechnica Sin, 2022, 53(4): 1041-1050. | |
| [96] |
王晶, 朱喆, 张鹏, 等. 利用单碱基编辑器定点突变猪肌肉生长抑制素基因的研究[J]. 中国畜牧兽医, 2022, 49(8): 2880-2887.
doi: 10.16431/j.cnki.1671-7236.2022.08.004 |
| Wang J, Zhu Z, Zhang P, et al. Site-directed mutagenesis of porcine myostatin gene using single base editor[J]. China Anim Husb Vet Med, 2022, 49(8): 2880-2887. | |
| [97] |
王煜, 宋瑞高, 赵建国, 等. 碱基编辑器介导的猪IGF2基因高效定点突变[J]. 中国畜牧兽医, 2020, 47(11): 3427-3435.
doi: 10.16431/j.cnki.1671-7236.2020.11.002 |
| Wang Y, Song RG, Zhao JG, et al. Efficient site-directed mutation of porcine IGF2 gene via base editors[J]. China Anim Husb Vet Med, 2020, 47(11): 3427-3435. | |
| [98] |
Song RG, Wang Y, Zheng QT, et al. One-step base editing in multiple genes by direct embryo injection for pig trait improvement[J]. Sci China Life Sci, 2022, 65(4): 739-752.
doi: 10.1007/s11427-021-2013-8 |
| [99] |
Li ZF, Duan XY, An XM, et al. Efficient RNA-guided base editing for disease modeling in pigs[J]. Cell Discov, 2018, 4: 64.
doi: 10.1038/s41421-018-0065-7 pmid: 30588328 |
| [100] |
Xie JK, Ge WK, Li N, et al. Efficient base editing for multiple genes and loci in pigs using base editors[J]. Nat Commun, 2019, 10(1): 2852.
doi: 10.1038/s41467-019-10421-8 pmid: 31253764 |
| [101] |
Zhu XX, Pan JS, Lin T, et al. Adenine base-editing-mediated exon skipping induces gene knockout in cultured pig cells[J]. Biotechnol Lett, 2022, 44(1): 59-76.
doi: 10.1007/s10529-021-03214-x |
| [102] |
Yuan HM, Yu TT, Wang LY, et al. Efficient base editing by RNA-guided cytidine base editors(CBEs)in pigs[J]. Cell Mol Life Sci, 2020, 77(4): 719-733.
doi: 10.1007/s00018-019-03205-2 |
| [103] |
Yao J, Wang Y, Cao CW, et al. CRISPR/Cas9-mediated correction of MITF homozygous point mutation in a Waardenburg syndrome 2A pig model[J]. Mol Ther Nucleic Acids, 2021, 24: 986-999.
doi: 10.1016/j.omtn.2021.04.009 URL |
| [104] |
Jiang HY, Jing QQ, Yang Q, et al. Efficient simultaneous introduction of premature stop codons in three tumor suppressor genes in PFFs via a cytosine base editor[J]. Genes, 2022, 13(5): 835.
doi: 10.3390/genes13050835 URL |
| [105] |
Zheng SW, Zhong HW, Zhou XQ, et al. Efficient and safe editing of porcine endogenous retrovirus genomes by multiple-site base-editing editor[J]. Cells, 2022, 11(24): 3975.
doi: 10.3390/cells11243975 URL |
| [106] | 宗媛, 高彩霞. 碱基编辑系统研究进展[J]. 遗传, 2019, 41(9): 777-800. |
| Zong Y, Gao CX. Progress on base editing systems[J]. Hereditas, 2019, 41(9): 777-800. | |
| [107] |
Arbab M, Shen MW, Mok B, et al. Determinants of base editing outcomes from target library analysis and machine learning[J]. Cell, 2020, 182(2): 463-480.e30.
doi: S0092-8674(20)30632-2 pmid: 32533916 |
| [108] |
Allen F, Crepaldi L, Alsinet C, et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks[J]. Nat Biotechnol, 2019, 37: 64-72.
doi: 10.1038/nbt.4317 URL |
| [1] | 张进威, 吴元霞, 孙静, 李晓开, 陆路, 李周权, 葛良鹏. 共生微生物对仔猪肠道发育、代谢和线粒体功能的影响[J]. 生物技术通报, 2024, 40(1): 332-343. |
| [2] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [3] | 石佳鑫, 刘凯, 朱金洁, 祁显涛, 谢传晓, 刘昌林. 基因编辑技术改良玉米株型增加杂交种产量[J]. 生物技术通报, 2023, 39(8): 62-69. |
| [4] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [5] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [6] | 吴莉丹, 冉雪琴, 牛熙, 黄世会, 李升, 王嘉福. 猪源致病性大肠杆菌基因组比较与毒力因子分析[J]. 生物技术通报, 2023, 39(12): 287-299. |
| [7] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [8] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [9] | 王松, 简晓平, 潘婉舒, 张永光, 王涛, 游玲. 玉米小曲酒糟发酵饲料对育肥猪肠道菌群的影响[J]. 生物技术通报, 2022, 38(9): 248-257. |
| [10] | 赖昕彤, 王柯岚, 由雨欣, 谭俊杰. 基于CRISPR/Cas系统的DNA碱基编辑研究进展[J]. 生物技术通报, 2022, 38(6): 1-12. |
| [11] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| [12] | 张豪, 李哲, 郭凯, 黄艳华, 郝永任. 绿色木霉Tv-1511组蛋白乙酰化酶编码基因TvGCN5的功能分析[J]. 生物技术通报, 2022, 38(5): 136-148. |
| [13] | 李虹仪, 彭国良, 肖正中, 张茂. 调控猪ETV5基因miRNA的筛选鉴定[J]. 生物技术通报, 2022, 38(5): 169-174. |
| [14] | 陈映丹, 张扬, 夏嫱, 孙虹霞. CRISPR/Cas基因编辑技术及其在微藻研究中的应用[J]. 生物技术通报, 2022, 38(5): 257-268. |
| [15] | 成温玉, 张博昕, 赵鸿远, 陈艳, 谢娟平. 天然产物抗猪流行性腹泻病毒研究进展[J]. 生物技术通报, 2022, 38(12): 127-136. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||