








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 253-261.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0301
姜宇舢1( ), 兰倩1, 王芳1, 姜亮1, 裴成成1,2(
), 兰倩1, 王芳1, 姜亮1, 裴成成1,2( )
)
收稿日期:2024-03-27
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
裴成成,女,博士,助理研究员,研究方向:作物遗传育种;E-mail: 2304609281@sxau.edu.cn作者简介:姜宇舢,女,硕士研究生,研究方向:作物学;E-mail: jiangyushan99@163.com
基金资助:
JIANG Yu-shan1( ), LAN Qian1, WANG Fang1, JIANG Liang1, PEI Cheng-cheng1,2(
), LAN Qian1, WANG Fang1, JIANG Liang1, PEI Cheng-cheng1,2( )
)
Received:2024-03-27
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】利用收集的藜麦植株颜色突变体,测定该突变体的代谢组成,分析代谢通路之间变化和转化规律,构建出该突变体代谢变化基本模型,为进一步鉴定和克隆影响藜麦重要代谢通路的关键遗传位点提供材料基础。【方法】利用正向遗传学手段,从藜麦常规品种‘藜红1号’(Red Quinoa 1, RQ1)后代中,筛选到一植株和穗部红色消退而绿色加深(green quinoa 1, gq1)的自然突变株作为材料,与其原始亲本RQ1相比较,利用非靶向代谢组学鉴定灌浆时期幼穗差异代谢成分,通过KEGG(Kyoto encyclopedia of genes and genomes)代谢途径分析和差异代谢物关联分析,揭示GQ1基因突变引发的关键代谢通路的变化。【结果】经过连续4个世代的遗传分析表明,gq1突变体植株颜色变异能够稳定遗传,且是由单个遗传位点控制。与其原始亲本RQ1相对比,在藜麦gq1突变体中检测到了409个差异代谢物,其中含量升高的代谢物有110个,299个代谢物含量降低。代谢组学分析发现,在藜麦gq1突变体中,对植物次级代谢有着重要影响的酪氨酸和以其为核心衍生出的其他代谢产物发生了整体性的降低。此外,包括6种人体必需氨基酸在内的多种氨基酸和TCA循环中的成分,在gq1突变体发生了显著的减少。【结论】通过对这些差异代谢进行KEGG代谢途径富集分析,表明GQ1基因突变造成了以酪氨酸为核心的初级代谢组分和次级代谢组分整体降低,意味着该基因可以成为协同优化藜麦初级代谢和次级代谢的关键遗传位点。
姜宇舢, 兰倩, 王芳, 姜亮, 裴成成. 一个影响酪氨酸代谢藜麦突变体的鉴定[J]. 生物技术通报, 2024, 40(10): 253-261.
JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism[J]. Biotechnology Bulletin, 2024, 40(10): 253-261.

图1 ‘藜红1号’(RQ1)及其突变体(gq1)生长发育各阶段的表型比较 A:2周时期幼苗;B:2周时期幼苗侧面;C:灌浆期植株;D:灌浆期幼穗;E:种子;比例尺:A, B, D, E = 1 cm, C = 10 cm
Fig. 1 Phenotypic comparison between ‘Red Quinoa 1’(RQ1)and its mutant(gq1)during different development stages A: 2-week-old seedling; B: side view of 2-week-old seedling; C: plants during the filling stage; D: young panicle of grain filling stage; E: seed. Scale bar: 1 cm for A, B, D, and E, respectively, and 10 cm for C
| 杂交组合 Hybrid combination of F2 | 总计 Total | 红色单株个体数 Number of red individuals | 绿色单株个体数 Number of green individuals | x2(3∶1) | P |
|---|---|---|---|---|---|
| RQ1/gq1 | 163 | 128 | 35 | 1.082 | 0.298 |
表1 突变体gq1和‘藜红1号’(RQ1)杂交F2世代性状分离比的卡方测验
Table 1 Test of Chi-square on segregation rate of F2 population between gq1 and RQ1
| 杂交组合 Hybrid combination of F2 | 总计 Total | 红色单株个体数 Number of red individuals | 绿色单株个体数 Number of green individuals | x2(3∶1) | P |
|---|---|---|---|---|---|
| RQ1/gq1 | 163 | 128 | 35 | 1.082 | 0.298 |
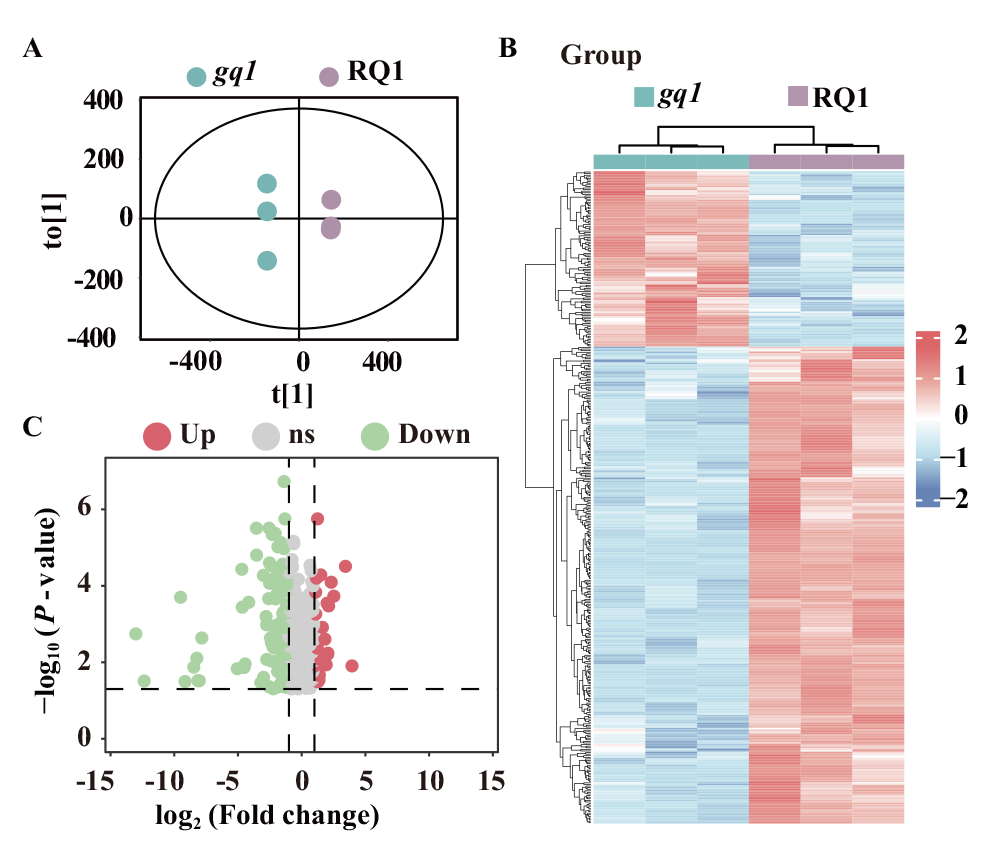
图2 ‘藜红1号’(RQ1)及其突变体(gq1)中差异代谢物分析概述 A:gq1对RQ1的OPLS-DA模型得分散点图;B:差异代谢物层次聚类分析热图;C:基于倍数变化的差异代谢物的火山图
Fig. 2 Overview of differential metabolite analysis in gq1 vs RQ1 A: Score plots of the OPLS-DA model for gq1 vs RQ1. B: Heatmap of hierarchical clustering analysis for differential metabolites. C: Volcano plot of differential metabolites based on fold change
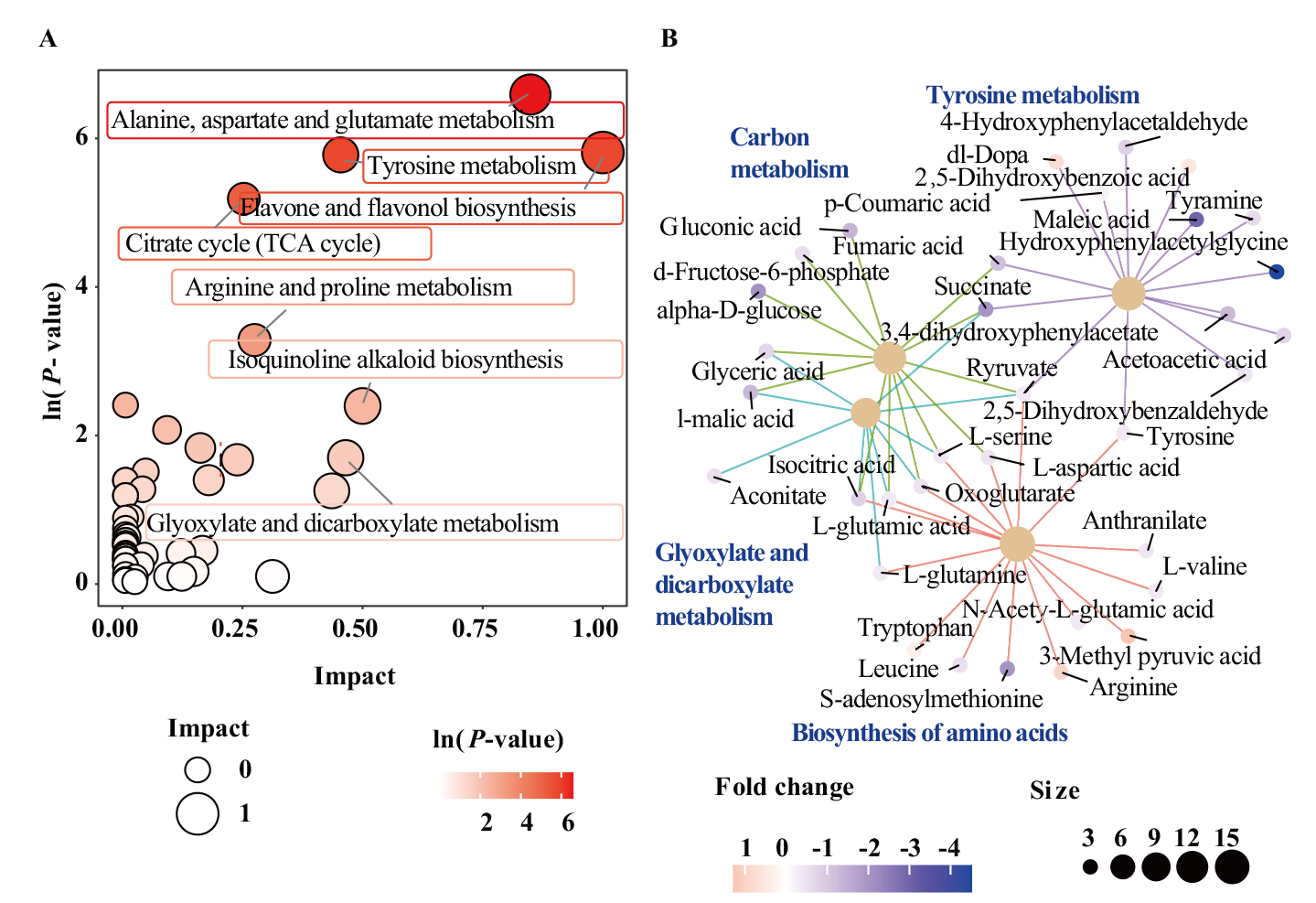
图4 ‘藜红1号’(RQ1)及其突变体(gq1)中差异代谢物KEGG通路分析 A:gq1组对RQ1组的代谢通路富集分析;B:gq1组对RQ1组的KEGG代谢互作分析
Fig. 4 KEGG pathway enrichment analysis of differential metabolites between RQ1 and gq1 A: KEGG pathway enrichment analysis of qg1 vs RQ1; B: KEGG category netplot of qg1 vs RQ1
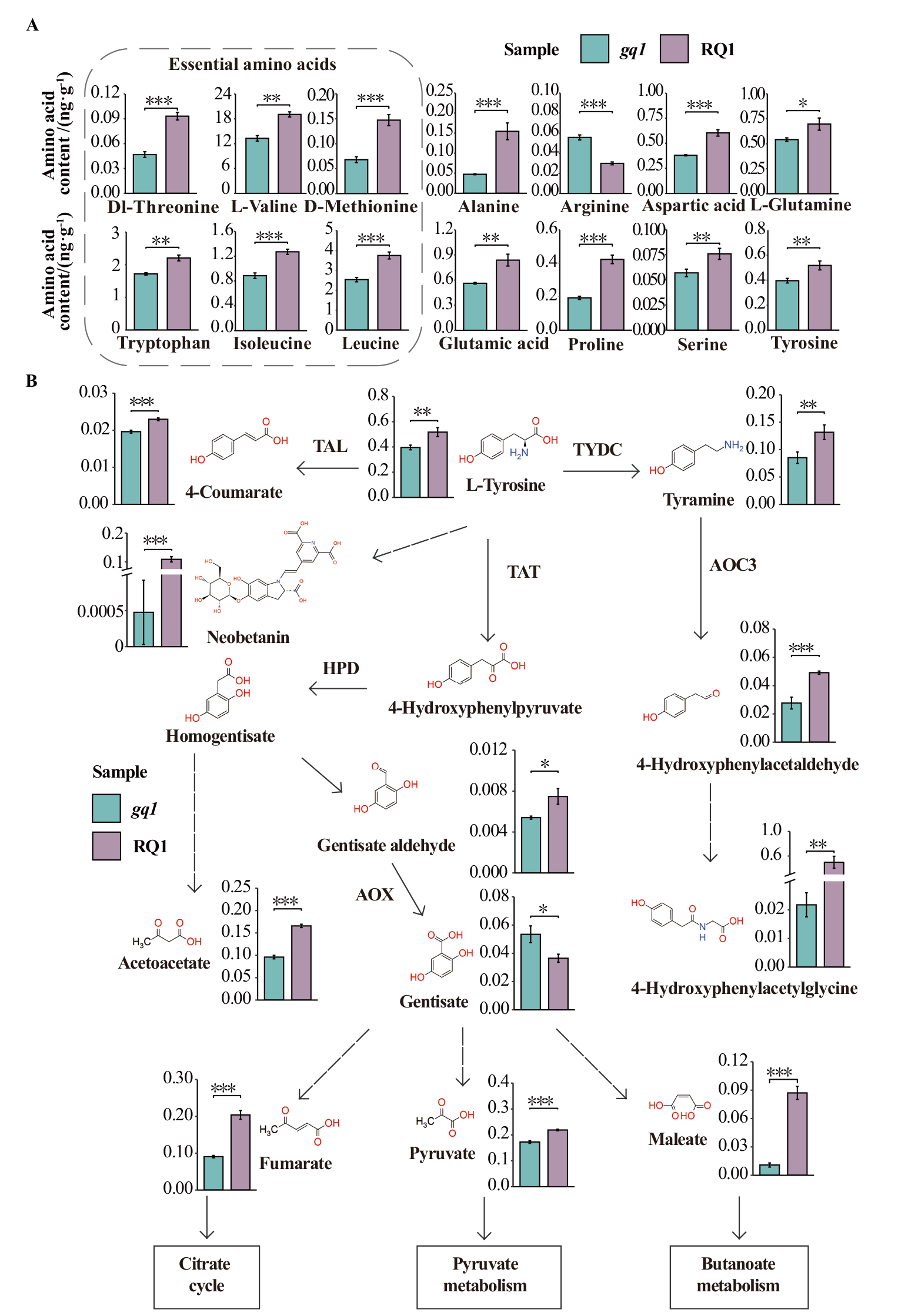
图5 gq1突变体中氨基酸的变化 A:gq1和RQ1中鉴定出的14种差异表达氨基酸含量;B:酪氨酸代谢途径中代谢物变化概述,虚线箭头表示多个酶步骤。TAT:酪氨酸转氨酶;TAL:酪氨酸解氨酶;TYDC:L-酪氨酸脱羧酶;HPD:羟苯丙酮酸双加氧酶;AOC3:伯胺氧化酶;AOX:醛氧化酶;差异的显著性(* P ≤ 0.05,** P ≤ 0.01,*** P ≤ 0.001)
Fig. 5 Variation of amino acid content in mutant gq1 A: Contents of 14 differentially expressed amino acids in gq1 and RQ1. B: An overview of metabolites and their expressions in tyrosine metabolism pathway. Arrows with dashed lines designate multiple enzymes steps. TAT: Tyrosine aminotransferase; TAL: tyrosine ammonia-lyase; TYDC: L-tyrosine decarboxylase; HPD: hydroxyphenylpyruvate dioxygenase; AOC3: primary-amine oxidase; AOX: aldehyde oxidase. Variables of significance(* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001)
| [1] | Alandia G, Rodriguez JP, Jacobsen SE, et al. Global expansion of quinoa and challenges for the Andean region[J]. Glob Food Secur, 2020, 26: 100429. |
| [2] | Abd El-Samad EH, Hussin SA, El-Naggar AM, et al. The potential use of quinoa as a new non-traditional leafy vegetable crop[J]. Bioscience Research, 2018, 15(4): 3387-3403. |
| [3] |
Filho AMM, Pirozi MR, Borges JT, et al. Quinoa: nutritional, functional, and antinutritional aspects[J]. Crit Rev Food Sci Nutr, 2017, 57(8): 1618-1630.
doi: 10.1080/10408398.2014.1001811 pmid: 26114306 |
| [4] |
Abugoch James LE. Quinoa(Chenopodium quinoa Willd.): composition, chemistry, nutritional, and functional properties[J]. Adv Food Nutr Res, 2009, 58: 1-31.
doi: 10.1016/S1043-4526(09)58001-1 pmid: 19878856 |
| [5] | Adolf VI, Shabala S, Andersen MN, et al. Varietal differences of quinoa's tolerance to saline conditions[J]. Plant Soil, 2012, 357(1): 117-129. |
| [6] | Sun WJ, Yao M, Wang Z, et al. Involvement of auxin-mediated CqEXPA50 contributes to salt tolerance in quinoa(Chenopodium quinoa)by interaction with auxin pathway genes[J]. Int J Mol Sci, 2022, 23(15): 8480. |
| [7] | Sun WJ, Wei JL, Wu GM, et al. CqZF-HD14 enhances drought tolerance in quinoa seedlings through interaction with CqHIPP34 and CqNAC79[J]. Plant Sci, 2022, 323: 111406. |
| [8] | Hinojosa L, González JA, Barrios-Masias FH, et al. Quinoa abiotic stress responses: a review[J]. Plants, 2018, 7(4): 106. |
| [9] | Jacobsen SE, Monteros C, Corcuera LJ, et al. Frost resistance mechanisms in quinoa(Chenopodium quinoa Willd.)[J]. Eur J Agron, 2007, 26(4): 471-475. |
| [10] | Ain QT, Siddique K, Bawazeer S, et al. Adaptive mechanisms in quinoa for coping in stressful environments: an update[J]. PeerJ, 2023, 11: e14832. |
| [11] | Graf BL, Rojas-Silva P, Rojo LE, et al. Innovations in health value and functional food development of quinoa(Chenopodium quinoa Willd.)[J]. Compr Rev Food Sci Food Saf, 2015, 14(4): 431-445. |
| [12] | Carciochi RA, Galván-D'Alessandro L, Vandendriessche P, et al. Effect of germination and fermentation process on the antioxidant compounds of quinoa seeds[J]. Plant Foods Hum Nutr, 2016, 71(4): 361-367. |
| [13] | Escuredo O, González Martín MI, Wells Moncada G, et al. Amino acid profile of the quinoa(Chenopodium quinoa Willd.) using near infrared spectroscopy and chemometric techniques[J]. J Cereal Sci, 2014, 60(1): 67-74. |
| [14] | Repo-Carrasco R, Espinoza C, Jacobsen SE. Nutritional value and use of the Andean crops quinoa(Chenopodium quinoa)and kañiwa(Chenopodium pallidicaule)[J]. Food Rev Int, 2003, 19(1/2): 179-189. |
| [15] | Guo HM, Hao YQ, Yang XS, et al. Exploration on bioactive properties of quinoa protein hydrolysate and peptides: a review[J]. Crit Rev Food Sci Nutr, 2023, 63(16): 2896-2909. |
| [16] |
Tang Y, Li XH, Chen PX, et al. Characterisation of fatty acid, carotenoid, tocopherol/tocotrienol compositions and antioxidant activities in seeds of three Chenopodium quinoa Willd. genotypes[J]. Food Chem, 2015, 174: 502-508.
doi: 10.1016/j.foodchem.2014.11.040 pmid: 25529712 |
| [17] | Chen X, Zhang YY, Cao BE, et al. Assessment and comparison of nutritional qualities of thirty quinoa(Chenopodium quinoa Willd.) seed varieties[J]. Food Chem X, 2023, 19: 100808. |
| [18] | Adamczewska-Sowińska K, Sowiński J, Jama-Rodzeńska A. The effect of sowing date and harvest time on leafy greens of quinoa(Chenopodium quinoa Willd.) yield and selected nutritional parameters[J]. Agriculture, 2021, 11(5): 405. |
| [19] | Pathan S, Eivazi F, Valliyodan B, et al. Nutritional composition of the green leaves of quinoa(Chenopodium quinoa Willd.)[J]. J Food Res, 2019, 8(6): 55. |
| [20] | Pathan S, Siddiqui RA. Nutritional composition and bioactive components in quinoa(Chenopodium quinoa Willd.) greens: a review[J]. Nutrients, 2022, 14(3): 558. |
| [21] | Qian GT, Li XY, Zhang H, et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa(Chenopodium quinoa Willd.) grains of different colors[J]. Food Chem X, 2023, 17: 100594. |
| [22] | Liu JN, Liu J, Zhang P, et al. Elucidating the differentiation synthesis mechanisms of differently colored resistance quinoa seedings using metabolite profiling and transcriptome analysis[J]. Metabolites, 2023, 13(10): 1065. |
| [23] | Liu YJ, Liu JN, Kong ZY, et al. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains[J]. Genomics, 2022, 114(1): 138-148. |
| [24] | Liu YJ, Liu JN, Li L, et al. Transcriptome and metabolome combined to analyze quinoa grain quality differences of different colors cultivars[J]. Int J Mol Sci, 2022, 23(21): 12883. |
| [25] | Cai YP, Weng K, Guo Y, et al. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing[J]. Metabolomics, 2015, 11(6): 1575-1586. |
| [26] |
Smith CA, Want EJ, O'Maille G, et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification[J]. Anal Chem, 2006, 78(3): 779-787.
doi: 10.1021/ac051437y pmid: 16448051 |
| [27] | Trygg J, Wold S. Orthogonal projections to latent structures(O-PLS)[J]. J Chemom, 2002, 16(3): 119-128. |
| [28] |
Gu ZG, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data[J]. Bioinformatics, 2016, 32(18): 2847-2849.
doi: 10.1093/bioinformatics/btw313 pmid: 27207943 |
| [29] | Wu TZ, Hu EQ, Xu SB, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data[J]. Innovation(Camb), 2021, 2(3): 100141. |
| [30] |
Galili G, Amir R, Fernie AR. The regulation of essential amino acid synthesis and accumulation in plants[J]. Annu Rev Plant Biol, 2016, 67: 153-178.
doi: 10.1146/annurev-arplant-043015-112213 pmid: 26735064 |
| [31] |
Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants[J]. Annu Rev Plant Biol, 2012, 63: 73-105.
doi: 10.1146/annurev-arplant-042811-105439 pmid: 22554242 |
| [32] |
Sandell FL, Holzweber T, Street NR, et al. Genomic basis of seed colour in quinoa inferred from variant patterns using extreme gradient boosting[J]. Plant Biotechnol J, 2024, 22(5): 1312-1324.
doi: 10.1111/pbi.14267 pmid: 38213076 |
| [1] | 林彤, 袁程, 董陈文华, 曾孟琼, 杨燕, 毛自朝, 林春. 藜麦配子发育相关基因CqSTK的筛选及功能分析[J]. 生物技术通报, 2024, 40(8): 83-94. |
| [2] | 孙慧琼, 张春来, 王锡亮, 徐宏申, 窦苗苗, 杨博慧, 柴文婷, 赵珊珊, 姜晓东. 藜麦FLS基因家族的鉴定、表达及DNA变异分析[J]. 生物技术通报, 2024, 40(7): 172-182. |
| [3] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [4] | 许沛冬, 易剑锋, 陈迪, 潘磊, 谢丙炎, 赵文军. 贝莱斯芽孢杆菌生防次级代谢产物研究进展[J]. 生物技术通报, 2024, 40(3): 75-88. |
| [5] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| [6] | 韩乐乐, 宋文迪, 边嘉珅, 李阳, 杨双胜, 陈紫怡, 李晓薇. 转录组与代谢组联合分析揭示大豆GmERD15c参与盐胁迫下类黄酮的生物合成[J]. 生物技术通报, 2024, 40(10): 243-252. |
| [7] | 何诗瑜, 曾仲大, 李博岩. 空间分辨代谢组学在疾病诊断研究中的应用进展[J]. 生物技术通报, 2024, 40(1): 145-159. |
| [8] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| [9] | 韩华蕊, 杨宇琭, 门艺涵, 韩尚玲, 韩渊怀, 霍轶琼, 侯思宇. 基于代谢组学研究谷子SiYABBYs参与花发育过程中鼠李糖苷的生物合成[J]. 生物技术通报, 2023, 39(6): 189-198. |
| [10] | 和梦颖, 刘文彬, 林震鸣, 黎尔彤, 汪洁, 金小宝. 一株抗革兰阳性菌的戈登氏菌WA4-43全基因组测序与分析[J]. 生物技术通报, 2023, 39(2): 232-242. |
| [11] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [12] | 古丽加马力·艾萨, 邢军, 李安, 张瑞. 开菲尔粒中微生物对苯并(α)芘的非靶向代谢组学分析[J]. 生物技术通报, 2022, 38(5): 123-135. |
| [13] | 杨玉萍, 张霞, 王翀翀, 王晓艳. 不同年龄大鼠尿液代谢组学研究[J]. 生物技术通报, 2022, 38(2): 166-172. |
| [14] | 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193. |
| [15] | 吴玉苹, 周勇, 蒲娟, 李会, 章金刚, 朱艳平. 代谢组学在肿瘤药物靶点筛选中的应用进展[J]. 生物技术通报, 2022, 38(1): 311-318. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||