








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 137-149.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0219
高萌萌1,2,3( ), 赵天宇1,2,3, 焦馨悦1,2,3, 林春晶2,3, 关哲允2,3, 丁孝羊2,3, 孙妍妍2,3(
), 赵天宇1,2,3, 焦馨悦1,2,3, 林春晶2,3, 关哲允2,3, 丁孝羊2,3, 孙妍妍2,3( ), 张春宝2,3(
), 张春宝2,3( )
)
收稿日期:2024-03-07
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
张春宝,男,博士,研究员,研究方向:大豆杂种优势利用及分子机理;E-mail: cbzhang@cjaas.com;作者简介:高萌萌,女,硕士研究生,研究方向:大豆杂种优势利用及分子机理;E-mail: 1757753098@qq.com
基金资助:
GAO Meng-meng1,2,3( ), ZHAO Tian-yu1,2,3, JIAO Xin-yue1,2,3, LIN Chun-jing2,3, GUAN Zhe-yun2,3, DING Xiao-yang2,3, SUN Yan-yan2,3(
), ZHAO Tian-yu1,2,3, JIAO Xin-yue1,2,3, LIN Chun-jing2,3, GUAN Zhe-yun2,3, DING Xiao-yang2,3, SUN Yan-yan2,3( ), ZHANG Chun-bao2,3(
), ZHANG Chun-bao2,3( )
)
Received:2024-03-07
Published:2024-07-26
Online:2024-07-30
摘要:
【目的】 “三系”法是选育杂交大豆的主要途径,但恢复系中仅有少数大豆恢复基因被克隆,为挖掘更多新恢复基因,从而促进多基因聚合的强恢复系选育,提高杂种F1的育性稳定性。【方法】 以大豆细胞质雄性不育系JLCMS5A和其恢复系JLR2为材料,采用转录组测序技术分析JLCMS5A和JLR2花蕾不同时期的转录水平变化,挖掘调控育性恢复和花蕾发育进程的相关基因和通路。进一步对具有恢复基因特征编码PPR(pentatricopeptide repeat)蛋白的差异表达基因(differentially expressed genes,DEGs)进行基因注释、序列差异分析、RT-qPCR验证、系统进化分析及蛋白结构预测,挖掘育性恢复相关基因。【结果】 转录组测序鉴定到17 181个DEGs,其中有3 856个DEGs与发育时期相关,2 808个DEGs与育性相关。GO(gene ontology)功能注释表明,与育性相关的DEGs主要富集在ADP结合、核酸结合转录因子活性和蛋白激酶活性等功能类别,与发育时期相关的DEGs主要富集在蛋白质异源二聚化活性、DNA复制和DNA结合等功能类别。KEGG(kyoto encyclopedia of genes and genomes)富集分析表明,育性相关DEGs主要参与内质网蛋白质加工、葡萄糖苷酸生物合成和植物激素信号转导等与蛋白质、糖类和信号转导密切相关的代谢通路,发育时期相关DEGs主要参与DNA复制、错配修复、淀粉和蔗糖代谢等与细胞分裂和能量物质降解密切相关的代谢通路。育性恢复候选基因分析发现,JLR2中的Glyma.09G176400可能在调控大豆细胞质雄性不育育性恢复过程中起到一定作用。【结论】 共鉴定到3 856个与发育时期相关的DEGs,2 808个与育性相关的DEGs;鉴定出具有恢复基因特征编码PPR蛋白的DEGs 15个;挖掘了1个育性恢复相关基因Glyma.09G176400。
高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149.
GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean[J]. Biotechnology Bulletin, 2024, 40(7): 137-149.
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Cons4 | GATCAGCAATTATGCACAACG | CCGCCACCATTCAGATTATGT |
| Glyma.01G235800 | CGTGAGAGATGTGGTGACATAC | ACGACCCATCTTCCATTTCC |
| Glyma.08G233900 | TGAGAGATGTGGTGCTTTGG | :GTTTCCCTCCATCCTCCTAAAC |
| Glyma.09G160300 | GATTGGTAGAAGAGCGACTGAA | CGACCGAGAAGATCGATCAAA |
| Glyma.09G176400 | GGTCAACCCGCAGATGTAAT | CGGTCGAATTCCCTGATCTTT |
| Glyma.09G176600 | GTCGCTGCAAACAAGGTTAAG | GGCTGAGAGTGAAAGTGAGAAT |
| Glyma.09G209700 | CAGGACAAGTCTCAACCACAA | CTGTGACATGTCTGGCGTATAA |
| Glyma.10G047150 | CTTCGTGAAGGAGAGTGGAATC | CAAACAGCTCCCGCGTATAA |
| Glyma.13G135000 | CGCGGGTTGTACTCTGATATT | GTTCCACGTTACGATATCTCTCTC |
| Glyma.15G019900 | TGAGCCGGATATTGTGGTTTAC | ACCTCTTCCTCCTCATCTCTTT |
| Glyma.15G091700 | GTTGTATGGGTGTGAGCCTAAT | CCTTCCCTCTCATCTCCCTATAA |
| Glyma.16G034600 | CCGAGGAGTTCAAGTCTTCTTC | CCTGGTTCTTTCGCTGGTAATA |
| Glyma.16G195700 | CTAGGAGATGCCTGTGATTTGT | AAAGCCATGGATCAGAGTAGTG |
| Glyma.16G195900 | CCGGATGCAATTACCCTCAA | ACCCTTGAGCTACGACCTTA |
| Glyma.17G220100 | GTGCCCATAGGAGTCAGAAATAC | CGCGGAGATTCTTCGTTACTT |
| Glyma.18G108202 | GCGGCTTGAGGAAGGTAATAA | TCATGCATCCAGCCTCTTAATC |
表1 RT-qPCR引物序列信息
Table 1 Information of RT-qPCR primer sequences
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Cons4 | GATCAGCAATTATGCACAACG | CCGCCACCATTCAGATTATGT |
| Glyma.01G235800 | CGTGAGAGATGTGGTGACATAC | ACGACCCATCTTCCATTTCC |
| Glyma.08G233900 | TGAGAGATGTGGTGCTTTGG | :GTTTCCCTCCATCCTCCTAAAC |
| Glyma.09G160300 | GATTGGTAGAAGAGCGACTGAA | CGACCGAGAAGATCGATCAAA |
| Glyma.09G176400 | GGTCAACCCGCAGATGTAAT | CGGTCGAATTCCCTGATCTTT |
| Glyma.09G176600 | GTCGCTGCAAACAAGGTTAAG | GGCTGAGAGTGAAAGTGAGAAT |
| Glyma.09G209700 | CAGGACAAGTCTCAACCACAA | CTGTGACATGTCTGGCGTATAA |
| Glyma.10G047150 | CTTCGTGAAGGAGAGTGGAATC | CAAACAGCTCCCGCGTATAA |
| Glyma.13G135000 | CGCGGGTTGTACTCTGATATT | GTTCCACGTTACGATATCTCTCTC |
| Glyma.15G019900 | TGAGCCGGATATTGTGGTTTAC | ACCTCTTCCTCCTCATCTCTTT |
| Glyma.15G091700 | GTTGTATGGGTGTGAGCCTAAT | CCTTCCCTCTCATCTCCCTATAA |
| Glyma.16G034600 | CCGAGGAGTTCAAGTCTTCTTC | CCTGGTTCTTTCGCTGGTAATA |
| Glyma.16G195700 | CTAGGAGATGCCTGTGATTTGT | AAAGCCATGGATCAGAGTAGTG |
| Glyma.16G195900 | CCGGATGCAATTACCCTCAA | ACCCTTGAGCTACGACCTTA |
| Glyma.17G220100 | GTGCCCATAGGAGTCAGAAATAC | CGCGGAGATTCTTCGTTACTT |
| Glyma.18G108202 | GCGGCTTGAGGAAGGTAATAA | TCATGCATCCAGCCTCTTAATC |
图2 花蕾总RNA琼脂糖凝胶电泳检测结果 M: Maker Ⅲ;1-3分别为FLB_1-FLB_3;4-6分别为FSB_1-FSB_3;7-9分别为SLB_1-SLB_3;10-12分别为SSB_1-SSB_3
Fig. 2 Result of total RNA in flower bud by agarose gel electrophoresis M: Maker Ⅲ. Lane 1-3: FLB_1 - FLB_3; respectively. Lane 4-6: FSB_1 - FSB_3; respectively. Lane 7-9: SLB_1 -SLB_3; respectively. Lane 10-12: SSB_1 - SSB_3; respectively
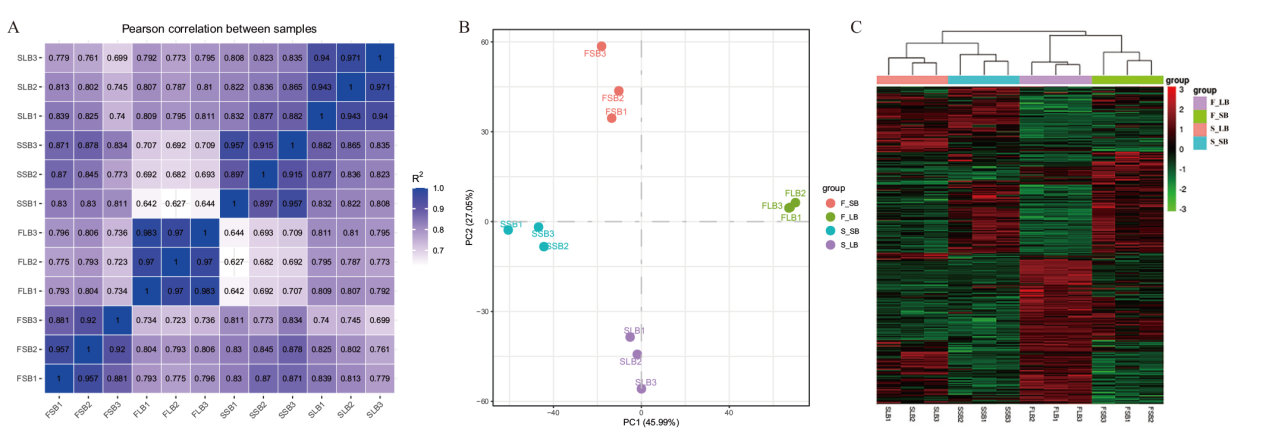
图3 样本间相关性检验 A:不同育性花蕾不同发育时期样本Pearson相关性分析;B:不同育性花蕾不同发育时期样本基因表达量PCA分析;C:样本间基因表达水平热图
Fig. 3 Correlation analysis among samples A: Pearson correlation analysis of fertile and sterile samples at different developmental stages. B: PCA analysis of gene expression levels in fertile and sterile samples at different developmental stages. C: Heat map of gene expressions among samples
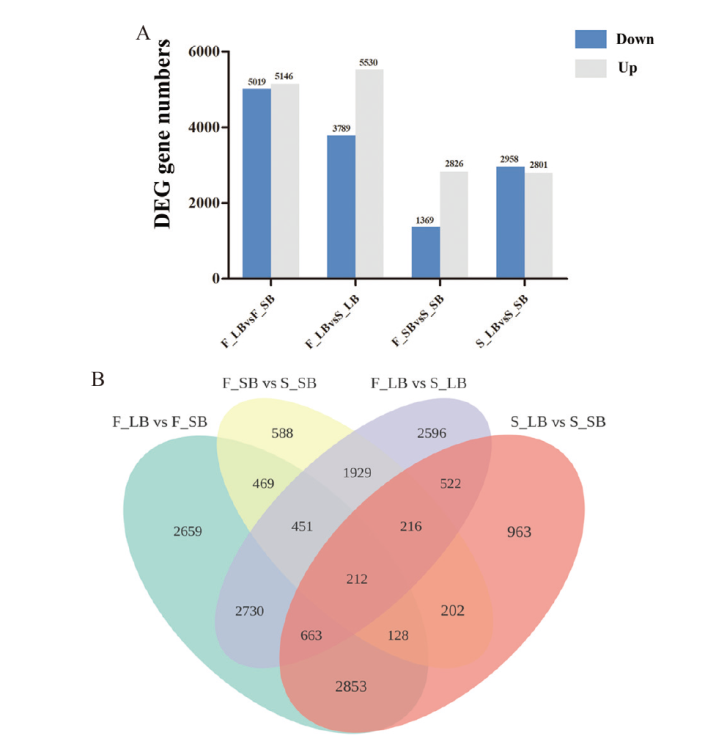
图4 差异表达基因的统计分析 A:差异表达基因数量统计;B:差异表达基因韦恩图
Fig. 4 Statistics and analysis of differentially expressed genes A: Statistics of differentially expressed genes. B: Venn diagram of differentially expressed genes
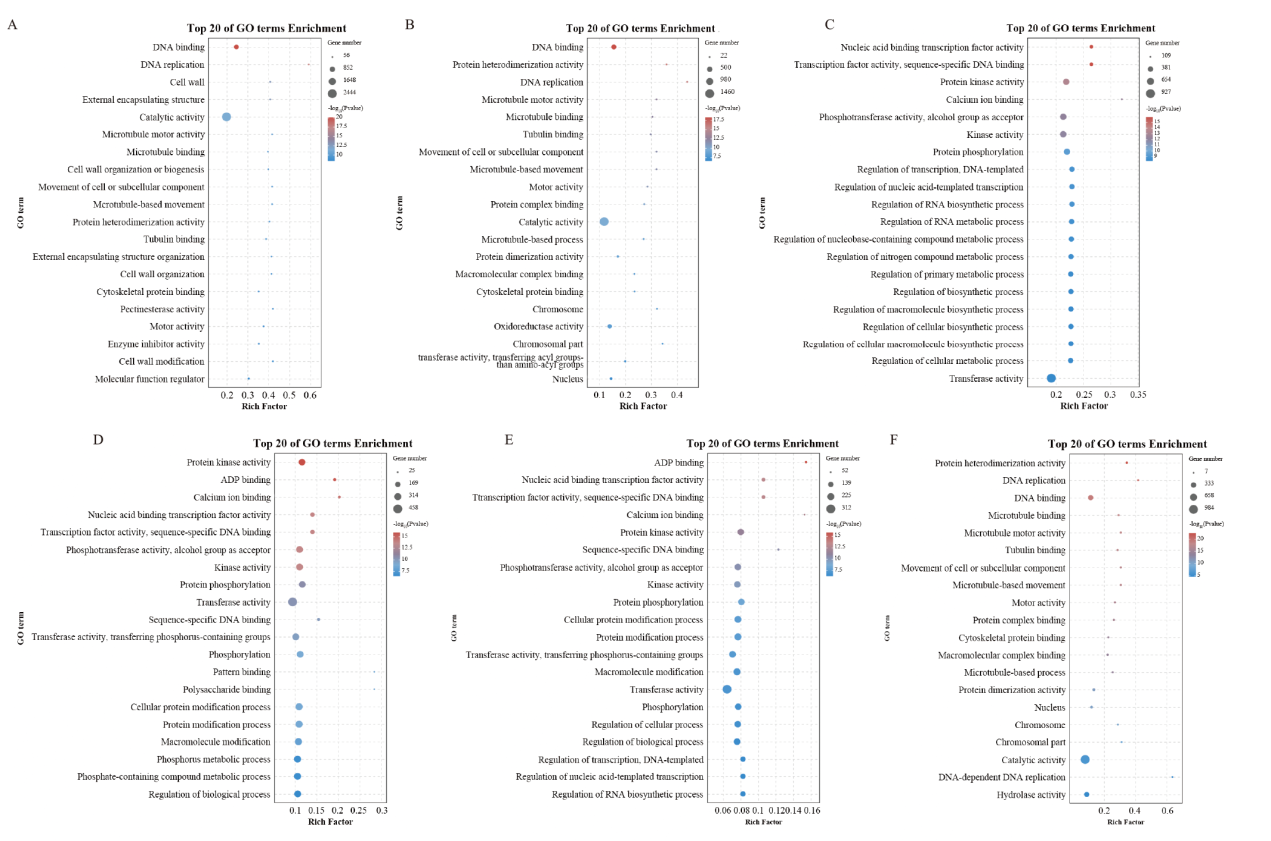
图5 GO富集分析散点图
Fig. 5 Rich distribution point diagram for GO A:F_LB vs F_SB;B:S_LB vs S_SB;C:F_LB vs S_LB;D:F_SB vs S_SB;E:F_LB vs S_LB和F_SB vs S_SB;F:F_LB vs F_SB和S_LB vs S_SB, the same below
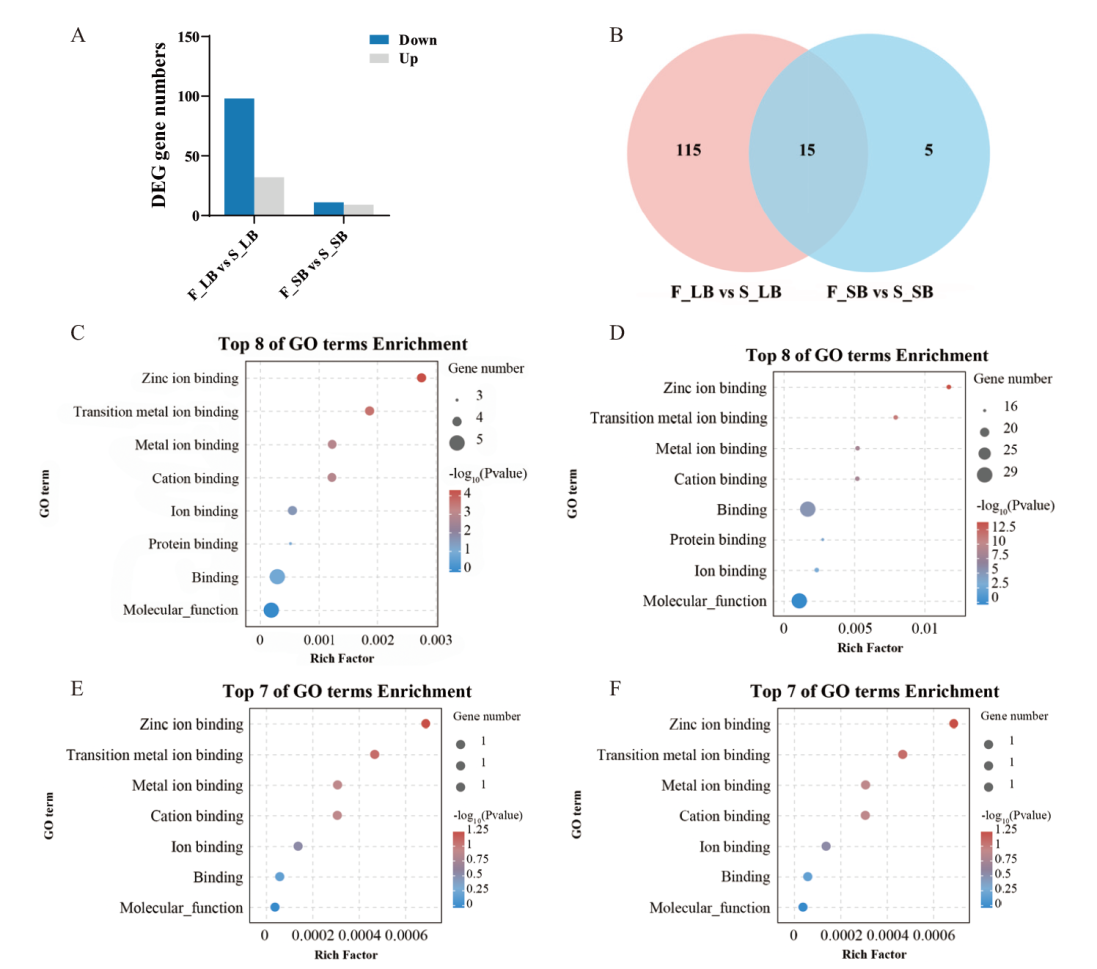
图7 编码PPR蛋白的差异表达基因统计分析 A:编码PPR蛋白的差异表达基因数量统计;B:编码PPR蛋白的差异表达基因Venn图;C:F_LB vs S_LB上调表达基因GO富集图;D:F_LB vs S_LB下调表达基因GO富集图;E:F_SB vs S_SB上调表达基因GO富集图;F:F_SB vs S_SB下调表达基因GO富集图
Fig. 7 Statistical analysis of differentially expressed genes encoding PPR proteins A: Statistics of differentially expressed genes encoding PPR protein. B: Venn diagram of differentially expressed genes encoding PPR protein. C: Map of GO enrichment of up-regulated DEGs in F_LB vs S_LB. D: Map of GO enrichment of down-regulated DEGs in F_LB vs S_LB. E: Map of GO enrichment of up-regulated DEGs in F_SB vs S_SB. F: Map of GO enrichment of down-regulated DEGs in F_SB vs S_SB
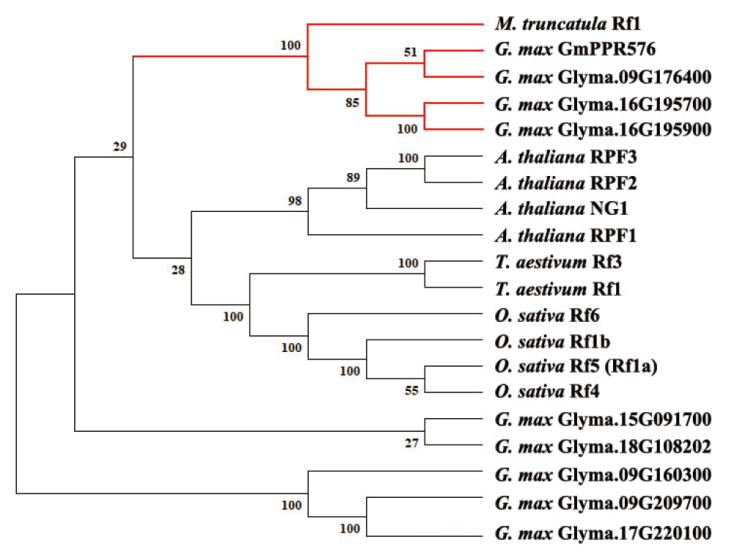
图9 候选基因编码PPR蛋白进化分析 红色分支代表豆科植物中的PPR蛋白;利用Maximum-Likelihood法构建进化树,设置Bootstrap为500次重复
Fig. 9 Phylogenetic analysis of candidate genes encoding PPR proteins The red branches indicate PPR proteins in leguminous plants. Phylogenetic analysis was constructed using Maximum-Likelihood, and Bootstrap values of 500 were used
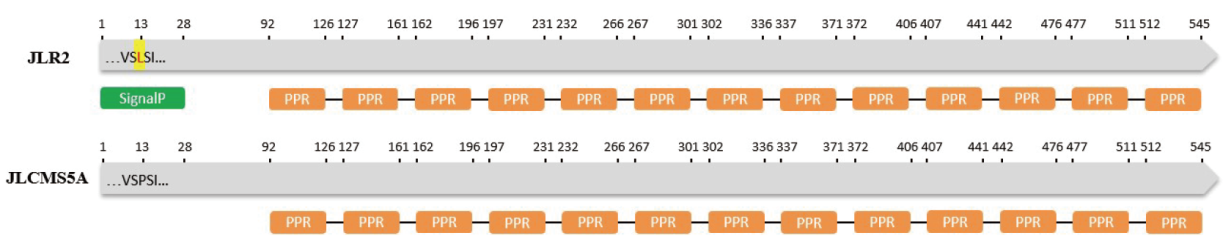
图10 Glyma.09G176400编码蛋白结构预测 绿色图形代表信号肽;橙色图形代表PPR,黄色图形代表氨基酸变异位点
Fig. 10 Structure prediction for Glyma.09G176400 encoded protein Green graphic indicates signal peptides. The orange graphics indicate pentatricopeptide repeat. The yellow graphic indicate amino acid mutation site
| [1] | 张春宝, 孙妍妍, 赵丽梅. 大豆细胞质雄性不育遗传基础与育种应用[J]. 植物遗传资源学报, 2024, 25(6): 857-869. |
| Zhang CB, Sun YY, Zhao LM. Genetic basis and breeding application of cytoplasmic male sterility in soybean[J]. J Plant Genet Resour, 2024, 25(6): 857-869. | |
| [2] |
Chen LT, Liu YG. Male sterility and fertility restoration in crops[J]. Annu Rev Plant Biol, 2014, 65: 579-606.
doi: 10.1146/annurev-arplant-050213-040119 pmid: 24313845 |
| [3] | Levings CS. Thoughts on cytoplasmic male sterility in cms-T maize[J]. Plant Cell, 1993, 5(10): 1285-1290. |
| [4] | Grelon M, Budar F, Bonhomme S, et al. Ogura cytoplasmic male-sterility(CMS)-associated orf138 is translated into a mitochondrial membrane polypeptide in male-sterile Brassica cybrids[J]. Mol Gen Genet, 1994, 243(5): 540-547. |
| [5] | Schnable P. The molecular basis of cytoplasmic male sterility and fertility restoration[J]. Trends Plant Sci, 1998, 3(5): 175-180. |
| [6] |
Balk J, Leaver CJ. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release[J]. Plant Cell, 2001, 13(8): 1803-1818.
doi: 10.1105/tpc.010116 pmid: 11487694 |
| [7] | Qin XE, Tian SK, Zhang WL, et al. The main restorer Rf3 of maize S type cytoplasmic male sterility encodes a PPR protein that functions in reduction of the transcripts of orf355[J]. Mol Plant, 2021, 14(12): 1961-1964. |
| [8] | Jiang HC, Lu Q, Qiu SQ, et al. Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice[J]. Proc Natl Acad Sci USA, 2022, 119(34): e2208759119. |
| [9] | Lin YN, Yang HL, Liu HM, et al. A P-type pentatricopeptide repeat protein ZmRF5 promotes 5' region partial cleavages of atp6c transcripts to restore the fertility of CMS-C maize by recruiting a splicing factor[J]. Plant Biotechnol J, 2023: 1-13. |
| [10] |
Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5' region of mRNAs whose translation it activates[J]. Plant Cell, 2005, 17(10): 2791-2804.
doi: 10.1105/tpc.105.034454 pmid: 16141451 |
| [11] |
Beick S, Schmitz-Linneweber C, Williams-Carrier R, et al. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts[J]. Mol Cell Biol, 2008, 28(17): 5337-5347.
doi: 10.1128/MCB.00563-08 pmid: 18591259 |
| [12] |
Pfalz J, Bayraktar OA, Prikryl J, et al. Site-specific binding of a PPR protein defines and stabilizes 5' and 3' mRNA termini in chloroplasts[J]. EMBO J, 2009, 28(14): 2042-2052.
doi: 10.1038/emboj.2009.121 pmid: 19424177 |
| [13] | Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts[J]. Nature, 2005, 433(7023): 326-330. |
| [14] | Okuda K, Hammani K, Tanz SK, et al. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts[J]. Plant J, 2010, 61(2): 339-349. |
| [15] |
Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript[J]. RNA, 2009, 15(6): 1142-1153.
doi: 10.1261/rna.1533909 pmid: 19395655 |
| [16] |
Gillman JD, Bentolila S, Hanson MR. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus[J]. Plant J, 2007, 49(2): 217-227.
doi: 10.1111/j.1365-313X.2006.02953.x pmid: 17156410 |
| [17] |
Brown GG, Formanová N, Jin H, et al. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats[J]. Plant J, 2003, 35(2): 262-272.
doi: 10.1046/j.1365-313x.2003.01799.x pmid: 12848830 |
| [18] |
Liu Z, Yang ZH, Wang X, et al. A mitochondria-targeted PPR protein restores pol cytoplasmic male sterility by reducing orf224 transcript levels in oilseed rape[J]. Mol Plant, 2016, 9(7): 1082-1084.
doi: 10.1016/j.molp.2016.04.004 pmid: 27102212 |
| [19] | Liu Z, Dong FM, Wang X, et al. A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus[J]. J Exp Bot, 2017, 68(15): 4115-4123. |
| [20] | Wang HD, Xiao Q, Wei C, et al. A mitochondria-localized pentatricopeptide repeat protein is required to restore hau cytoplasmic male sterility in Brassica napus[J]. Theor Appl Genet, 2021, 134(5): 1377-1386. |
| [21] | Komori T, Ohta S, Murai N, et al. Map-based cloning of a fertility restorer gene, Rf-1, in rice(Oryza sativa L.)[J]. Plant J, 2004, 37(3): 315-325. |
| [22] | Hu J, Wang K, Huang WC, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162[J]. Plant Cell, 2012, 24(1): 109-122. |
| [23] |
Tang HW, Luo DP, Zhou DG, et al. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts[J]. Mol Plant, 2014, 7(9): 1497-1500.
doi: S1674-2052(14)60953-9 pmid: 24728538 |
| [24] |
Huang WC, Yu CC, Hu J, et al. Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility[J]. Proc Natl Acad Sci USA, 2015, 112(48): 14984-14989.
doi: 10.1073/pnas.1511748112 pmid: 26578814 |
| [25] | Melonek J, Duarte J, Martin J, et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat[J]. Nat Commun, 2021, 12(1): 1036. |
| [26] |
Wang TL, He TT, Ding XL, et al. Confirmation of GmPPR576 as a fertility restorer gene of cytoplasmic male sterility in soybean[J]. J Exp Bot, 2021, 72(22): 7729-7742.
doi: 10.1093/jxb/erab382 pmid: 34397079 |
| [27] |
杨绪磊, 郭凤兰, 高萌萌, 等. 大豆CMS-RN型不育系育性恢复基因GmRf1的初步鉴定及其分子标记开发[J]. 植物遗传资源学报, 2023, 24(4): 1186-1193.
doi: 10.13430/j.cnki.jpgr.20221124001 |
| Yang XL, Guo FL, Gao MM, et al. Preliminary identification and molecular marker development of the restorer-of-fertility gene GmRf1 of CMS-RN type sterile lines in soybean[J]. J Plant Genet Resour, 2023, 24(4): 1186-1193. | |
| [28] |
Liu YC, Du HL, Li PC, et al. Pan-genome of wild and cultivated soybeans[J]. Cell, 2020, 182(1): 162-176.e13.
doi: S0092-8674(20)30618-8 pmid: 32553274 |
| [29] |
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930.
doi: 10.1093/bioinformatics/btt656 pmid: 24227677 |
| [30] |
Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation[J]. Nat Biotechnol, 2010, 28(5): 511-515.
doi: 10.1038/nbt.1621 pmid: 20436464 |
| [31] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| [32] |
Gaborieau L, Brown GG, Mireau H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility[J]. Front Plant Sci, 2016, 7: 1816.
doi: 10.3389/fpls.2016.01816 pmid: 27999582 |
| [33] | Li JJ, Yang SP, Gai JY. Transcriptome comparative analysis between the cytoplasmic male sterile line and fertile line in soybean(Glycine max(L.)Merr.)[J]. Genes Genom, 2017, 39(10): 1117-1127. |
| [34] | Li JJ, Han SH, Ding XL, et al. Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean(Glycine max(L.)Merr.)[J]. PLoS One, 2015, 10(5): e0126771. |
| [35] | 赵颖. 联合RNA-seq和iTRAQ技术挖掘大豆细胞质雄性不育相关基因[D]. 通辽: 内蒙古民族大学, 2023. |
| Zhao Y. Combined RNA-seq and iTRAQ technology to mine genes related to cytoplasmic male sterility in soybean[D]. Tongliao: Inner Mongolia University for the Nationalities, 2023. | |
| [36] | 姜童, 付翔, 王辉, 等. 簇生朝天椒雄性不育系及保持系的花器官及生理特性研究[J]. 北方园艺, 2018(11): 22-26. |
| Jiang T, Fu X, Wang H, et al. Flower morphological and physiological characters between male sterile line and maintainer line in cluster pepper[J]. North Hortic, 2018(11): 22-26. | |
| [37] | 鲁美宏, 孙万仓, 孔德晶, 等. 白菜型冬油菜不育系LRCMS花器生理生化特性及其雄蕊发育特征研究[J]. 西北植物学报, 2014, 34(3): 509-515. |
| Lu MH, Sun WC, Kong DJ, et al. Physiobiochemical characteristics and stamen development characteristics of LRCMS flower in winter rapeseed(Brassica campestris)[J]. Acta Bot Boreali Occidentalia Sin, 2014, 34(3): 509-515. | |
| [38] |
白志元, 杨玉花, 张瑞军. 不同恢复型大豆细胞质雄性不育杂种F1的转录组分析[J]. 植物遗传资源学报, 2022, 23(6): 1847-1855.
doi: 10.13430/j.cnki.jpgr.20220510001 |
| Bai ZY, Yang YH, Zhang RJ. Transcriptomic analysis of soybean cytoplasmic male sterile F1 hybrids from pollination with different restorer types[J]. J Plant Genet Resour, 2022, 23(6): 1847-1855. | |
| [39] | Barkan A, Rojas M, Fujii S, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins[J]. PLoS Genet, 2012, 8(8): e1002910. |
| [40] | Kazama T, Toriyama K. A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein[J]. Rice, 2014, 7(1): 28. |
| [41] |
Wang ZH, Zou YJ, Li XY, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing[J]. Plant Cell, 2006, 18(3): 676-687.
doi: 10.1105/tpc.105.038240 pmid: 16489123 |
| [42] | Wang CD, Lezhneva L, Arnal N, et al. The radish Ogura fertility restorer impedes translation elongation along its cognate CMS-causing mRNA[J]. Proc Natl Acad Sci USA, 2021, 118(35): e2105274118. |
| [1] | 廖杨梅, 赵国春, 翁学煌, 贾黎明, 陈仲. 无患子雄性不育品种‘琦蕊’不同发育时期雄花转录组分析[J]. 生物技术通报, 2024, 40(7): 197-206. |
| [2] | 王芳, 于璐, 齐泽铮, 周长军, 于吉东. 大豆镰刀菌根腐病拮抗菌的筛选及生防效果[J]. 生物技术通报, 2024, 40(7): 216-225. |
| [3] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [4] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [5] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [6] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [7] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [8] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [9] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [10] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [11] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [12] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [13] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [14] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [15] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||