








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 237-247.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1099
蒋天威1( ), 马培杰2, 李亚娇2, 陈才俊2, 刘晓霞2, 王小利2(
), 马培杰2, 李亚娇2, 陈才俊2, 刘晓霞2, 王小利2( )
)
收稿日期:2024-11-11
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
王小利,男,博士,研究员,研究方向 :分子植物学;E-mail: WangXiaoli_GIP@163.com作者简介:蒋天威,男,硕士研究生,研究方向 :分子植物育种;E-mail: JiangTianwei_GIP@163.com
基金资助:
JIANG Tian-wei1( ), MA Pei-jie2, LI Ya-jiao2, CHEN Cai-jun2, LIU Xiao-xia2, WANG Xiao-li2(
), MA Pei-jie2, LI Ya-jiao2, CHEN Cai-jun2, LIU Xiao-xia2, WANG Xiao-li2( )
)
Received:2024-11-11
Published:2025-07-26
Online:2025-07-22
摘要:
目的 分析不同光周期对二穗短柄草(Brachypodium distachyon,
蒋天威, 马培杰, 李亚娇, 陈才俊, 刘晓霞, 王小利. 二穗短柄草对光周期的代谢响应分析[J]. 生物技术通报, 2025, 41(7): 237-247.
JIANG Tian-wei, MA Pei-jie, LI Ya-jiao, CHEN Cai-jun, LIU Xiao-xia, WANG Xiao-li. Metabolic Response Analysis of Brachypodium distachyon to Photoperiods[J]. Biotechnology Bulletin, 2025, 41(7): 237-247.

图1 代谢物分类饼图及OPLS-DA得分图、总积分统计图A:739种代谢物分类饼图;B:5个样本差异的OPLS-DA得分图;C:长短日照样本间差异的OPLS-DA得分图;D:总积分和统计图。图B、C横坐标代表第一主成分得分(PC1),用于区分不同样本组,距离越远差异越大;纵坐标代表第一正交主成分得分(OC1),用于揭示组内的相关性,重复间距离越小,重复性越好。图B、D中LS0为ZT0时的样本,L12、L24为长日照处理12 h、24 h的样本,S12、S24为短日照处理12 h、24 h的样本。图C中LS0为对照,LD表示L12、L24合并为一组,SD表示S12、S24合并为一组。图D中方差分析使用Tukey多重比较方法,字母间的差异表示差异显著(P<0.05)。下同
Fig. 1 Pie chart of metabolite classification, OPLS-DA score plot, and total score statisticsA: Pie chart showing the classification of 739 metabolites. B: OPLS-DA score plot of the five sample groups showing differences. C: OPLS-DA score plot revealing differences between long-day and short-day samples. D: Statistical plot of the total sum of integrals. In Fig. B and C, the X-axis refers to the first principal component score (PC1), which distinguishes different sample groups. The greater the distance, the larger the difference between the groups. The Y-axis refers to the first orthogonal component score (OC1), which reveals the internal correlations of the groups. The smaller the distance between replicates, the higher the reproducibility. In Fig. B and D, LS0 refers to the samples at ZT0, L12 and L24 refer to the samples subjected to long-day treatment for 12 and 24 h, respectively, while S12 and S24 refers to the samples subjected to short-day treatment for 12 and 24 h, respectively. In Fig. C, LS0 is the control group, LD refers to the combined L12 and L24 groups, and SD refers to the combined S12 and S24 groups. In Fig. D, variance analysis was performed using the Tukey multiple comparisons method. Significant differences are indicated by letters, where different letters denote a statistically significant difference (P<0.05). The same below
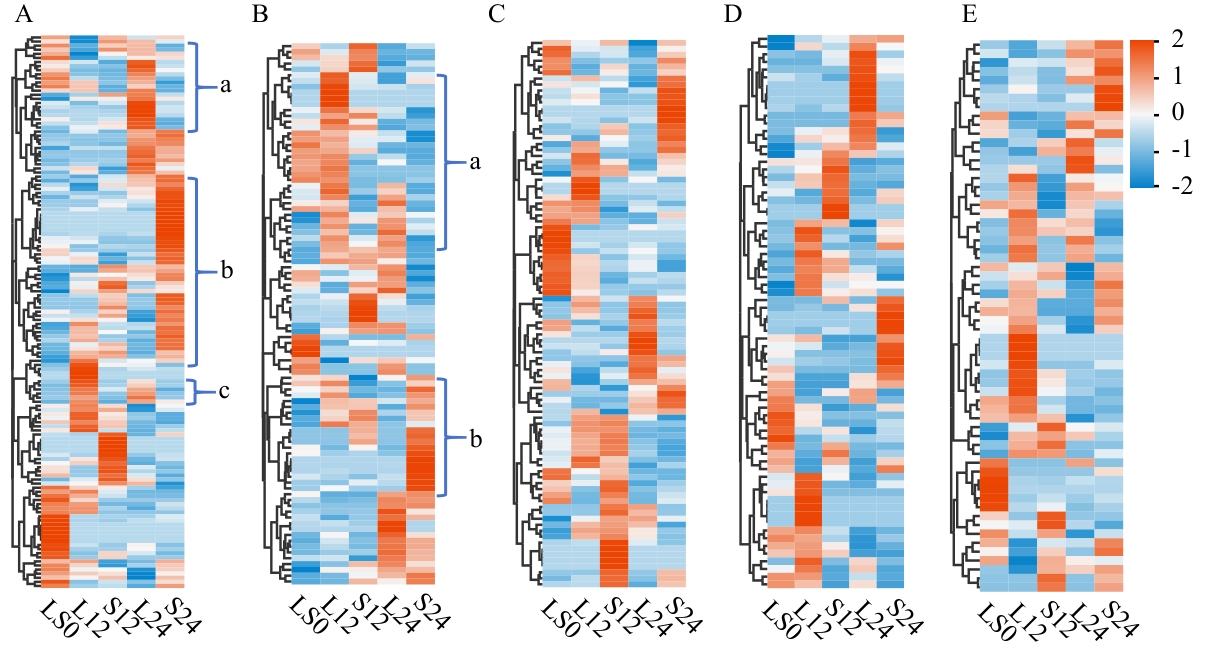
图2 五类代谢物聚类热图A:有机酸及其衍生物;B:有机含氧化合物;C:脂质和类脂分子;D:苯丙类和聚酮类;E:苯并杂环化合物。每个图均为行标准化、行聚类;红蓝分别代表上调及下调,小写字母a、b、c表示代谢物在长短日照间出现明显表达差异的区域
Fig. 2 Clustering heatmap of five classes of metabolitesA: Organic acids and their derivatives. B: Organic oxygen-containing compounds. C: Lipids and lipophilic molecules. D: Phenylpropanoids and polyketides. E: Benzoheterocyclic compounds. Each plot is row-normalized and row-clustered, with red and blue representing upregulation and downregulation, respectively. Lowercase letters a, b, and c indicate regions where metabolites show significant expression differences between long-day and short-day conditions
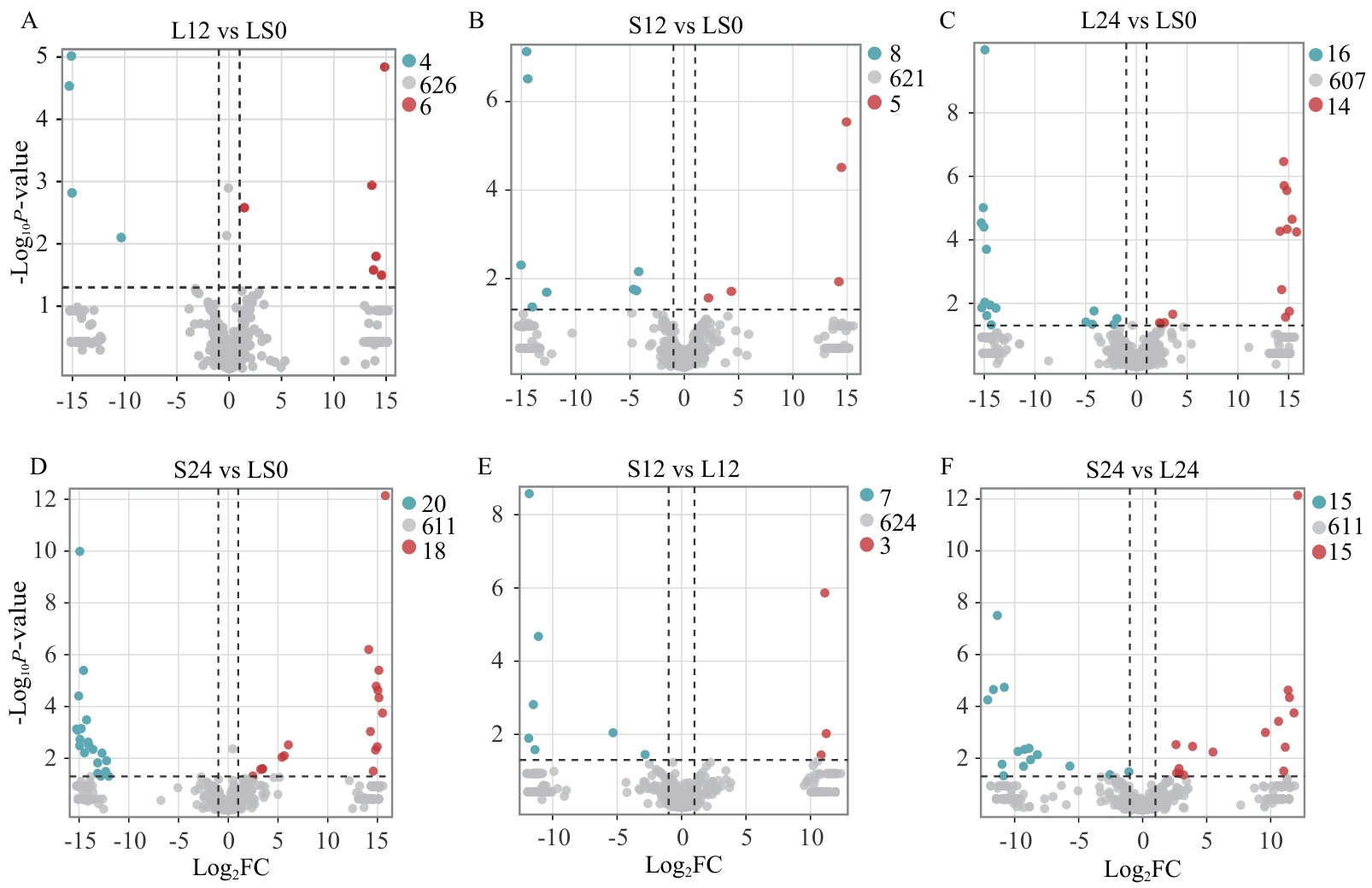
图3 各比较组差异代谢物火山图横坐标代表该组对比各物质的倍数变化(取以2为底的对数),纵坐标表示t检验的P值(取以10为底的对数)。点表示代谢物,红色、蓝色、灰色分别代表显著上调、显著下调、差异不显著,右上角点和数值表示不同颜色标注的代谢物数量
Fig. 3 Volcano plots of differential metabolites for each comparison groupThe x-axis refers to the fold change of each substance in the group comparison, taken as the logarithm base 2. The y-axis indicates the P-value from the t-test, taken as the logarithm base 10. Each dot refers to a metabolite, with red, blue, and gray colors corresponding to significantly up-regulated, significantly down-regulated, and non-significantly changed metabolites, respectively. The dots and numbers in the upper right corner indicate the number of metabolites labeled in different colors
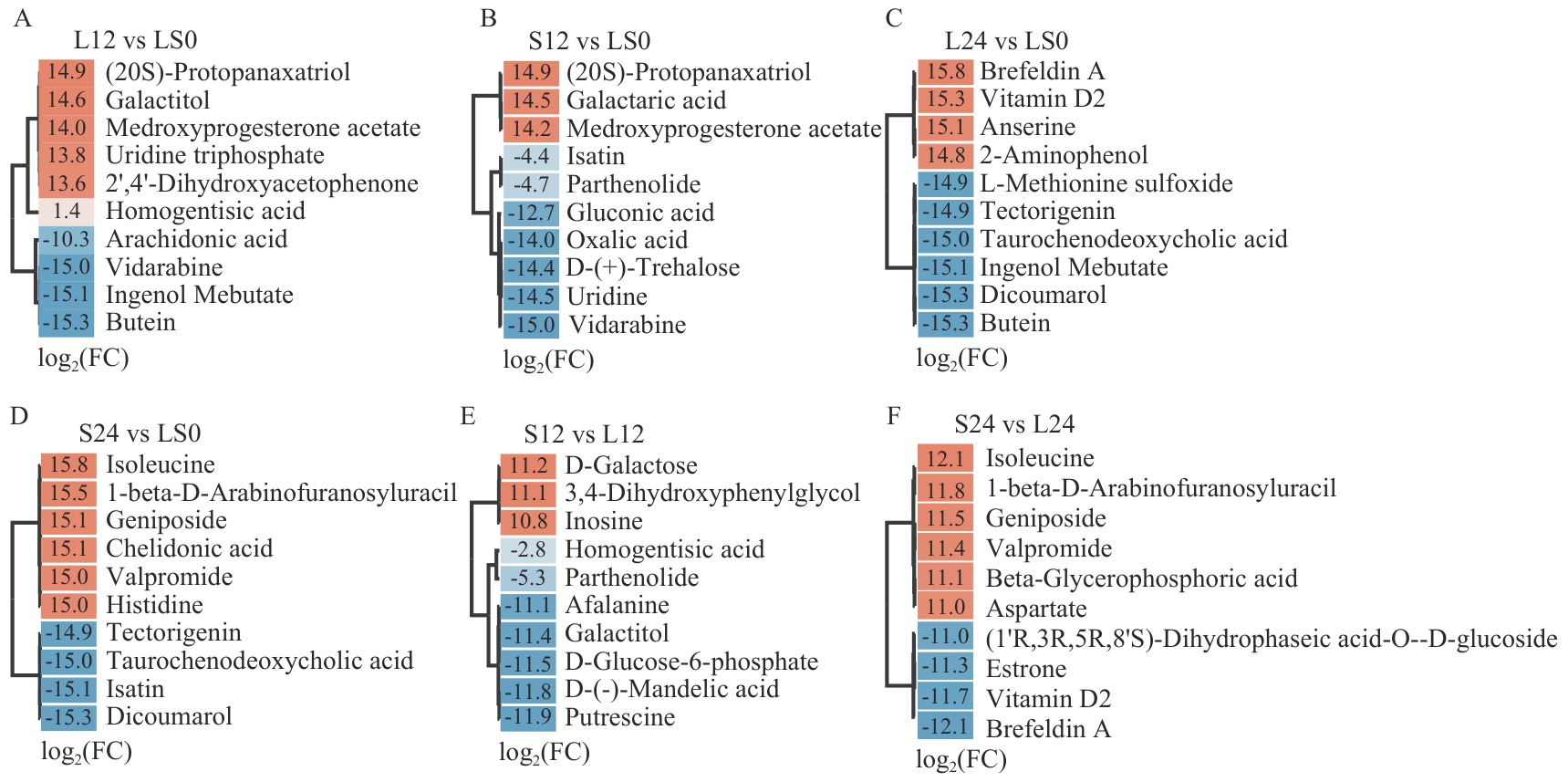
图4 各比较组中差异倍数最大的前10种DAM从浅色到深色的颜色梯度表示|log2FC|值增加,橙色表示上调,蓝色表示下调。标签显示|log2FC|值
Fig. 4 Top 10 DAMs with the highest fold changes in each comparison groupThe color gradient from light to dark indicates increasing |log2FC| values, with orange for up-regulation and blue for down-regulation. Labels show the |log2FC| values
组合 Group | 代谢物名称 Metabolite | VIP | Log2 倍变比 Log2(FC) | P值 P-value | 上调或下调 Up or down |
|---|---|---|---|---|---|
| S12 vs L12 | 腐胺 Putrescine | 2.08 | 11.86 | 1.28E-02 | DOWN |
| D-(-)-扁桃酸 D-(-)-mandelic acid | 2.30 | 11.82 | 2.65E-09 | DOWN | |
| D-葡萄糖-6-磷酸 D-glucose-6-phosphate | 2.23 | 11.50 | 1.54E-03 | DOWN | |
| 半乳糖醇 Galactitol | 1.98 | 11.37 | 2.62E-02 | DOWN | |
| D-半乳糖 D-galactose | 2.12 | 11.22 | 9.51E-03 | UP | |
| 3,4-二羟基苯乙二醇3,4-dihydroxyphenylglycol | 2.30 | 11.11 | 1.37E-06 | UP | |
| α-丙氨酸 Afalanine | 2.30 | 11.10 | 2.10E-05 | DOWN | |
| 次黄嘌呤核苷 Inosine | 1.94 | 10.83 | 3.65E-02 | UP | |
| 小白菊内酯 Parthenolide | 2.12 | 5.31 | 9.01E-03 | DOWN | |
| 尿黑酸 Homogentisic acid | 1.93 | 2.83 | 3.56E-02 | DOWN | |
| S24 vs L24 | 异亮氨酸 Isoleucine | 2.12 | 12.11 | 7.21E-13 | UP |
| 不枯芽菌素 A Brefeldin A | 2.11 | 12.10 | 5.61E-05 | DOWN | |
| 1-β-D-阿拉伯呋喃糖基尿嘧啶 1-beta-D-arabinofuranosyluracil | 2.10 | 11.82 | 1.79E-04 | UP | |
| 维生素 D2 Vitamin D2 | 2.11 | 11.66 | 2.24E-05 | DOWN | |
| 京尼平苷 Geniposide | 2.11 | 11.47 | 4.54E-05 | UP | |
| 丙戊酰胺 Valpromide | 2.11 | 11.36 | 2.37E-05 | UP | |
| 雌酮 Estrone | 2.12 | 11.35 | 3.09E-08 | DOWN | |
| β-甘油磷酸 Beta-Glycerophosphoric acid | 2.01 | 11.13 | 3.74E-03 | UP | |
| 天冬氨酸 Aspartate | 1.80 | 11.02 | 3.14E-02 | UP | |
| (1'R,3R,5R,8'S)-二氢相酸-O-β-D-葡萄糖苷 (1'R,3R,5R,8'S)-dihydrophaseic acid-O-D-glucoside | 1.89 | 10.99 | 1.71E-02 | DOWN |
表1 ZT12和ZT24二穗短柄草在SD和LD间差异显著的前10种DAM
Table 1 Top 10 DAM with significant differences between SD and LD conditions at ZT12 and ZT24 in Brachypodium distachyon
组合 Group | 代谢物名称 Metabolite | VIP | Log2 倍变比 Log2(FC) | P值 P-value | 上调或下调 Up or down |
|---|---|---|---|---|---|
| S12 vs L12 | 腐胺 Putrescine | 2.08 | 11.86 | 1.28E-02 | DOWN |
| D-(-)-扁桃酸 D-(-)-mandelic acid | 2.30 | 11.82 | 2.65E-09 | DOWN | |
| D-葡萄糖-6-磷酸 D-glucose-6-phosphate | 2.23 | 11.50 | 1.54E-03 | DOWN | |
| 半乳糖醇 Galactitol | 1.98 | 11.37 | 2.62E-02 | DOWN | |
| D-半乳糖 D-galactose | 2.12 | 11.22 | 9.51E-03 | UP | |
| 3,4-二羟基苯乙二醇3,4-dihydroxyphenylglycol | 2.30 | 11.11 | 1.37E-06 | UP | |
| α-丙氨酸 Afalanine | 2.30 | 11.10 | 2.10E-05 | DOWN | |
| 次黄嘌呤核苷 Inosine | 1.94 | 10.83 | 3.65E-02 | UP | |
| 小白菊内酯 Parthenolide | 2.12 | 5.31 | 9.01E-03 | DOWN | |
| 尿黑酸 Homogentisic acid | 1.93 | 2.83 | 3.56E-02 | DOWN | |
| S24 vs L24 | 异亮氨酸 Isoleucine | 2.12 | 12.11 | 7.21E-13 | UP |
| 不枯芽菌素 A Brefeldin A | 2.11 | 12.10 | 5.61E-05 | DOWN | |
| 1-β-D-阿拉伯呋喃糖基尿嘧啶 1-beta-D-arabinofuranosyluracil | 2.10 | 11.82 | 1.79E-04 | UP | |
| 维生素 D2 Vitamin D2 | 2.11 | 11.66 | 2.24E-05 | DOWN | |
| 京尼平苷 Geniposide | 2.11 | 11.47 | 4.54E-05 | UP | |
| 丙戊酰胺 Valpromide | 2.11 | 11.36 | 2.37E-05 | UP | |
| 雌酮 Estrone | 2.12 | 11.35 | 3.09E-08 | DOWN | |
| β-甘油磷酸 Beta-Glycerophosphoric acid | 2.01 | 11.13 | 3.74E-03 | UP | |
| 天冬氨酸 Aspartate | 1.80 | 11.02 | 3.14E-02 | UP | |
| (1'R,3R,5R,8'S)-二氢相酸-O-β-D-葡萄糖苷 (1'R,3R,5R,8'S)-dihydrophaseic acid-O-D-glucoside | 1.89 | 10.99 | 1.71E-02 | DOWN |
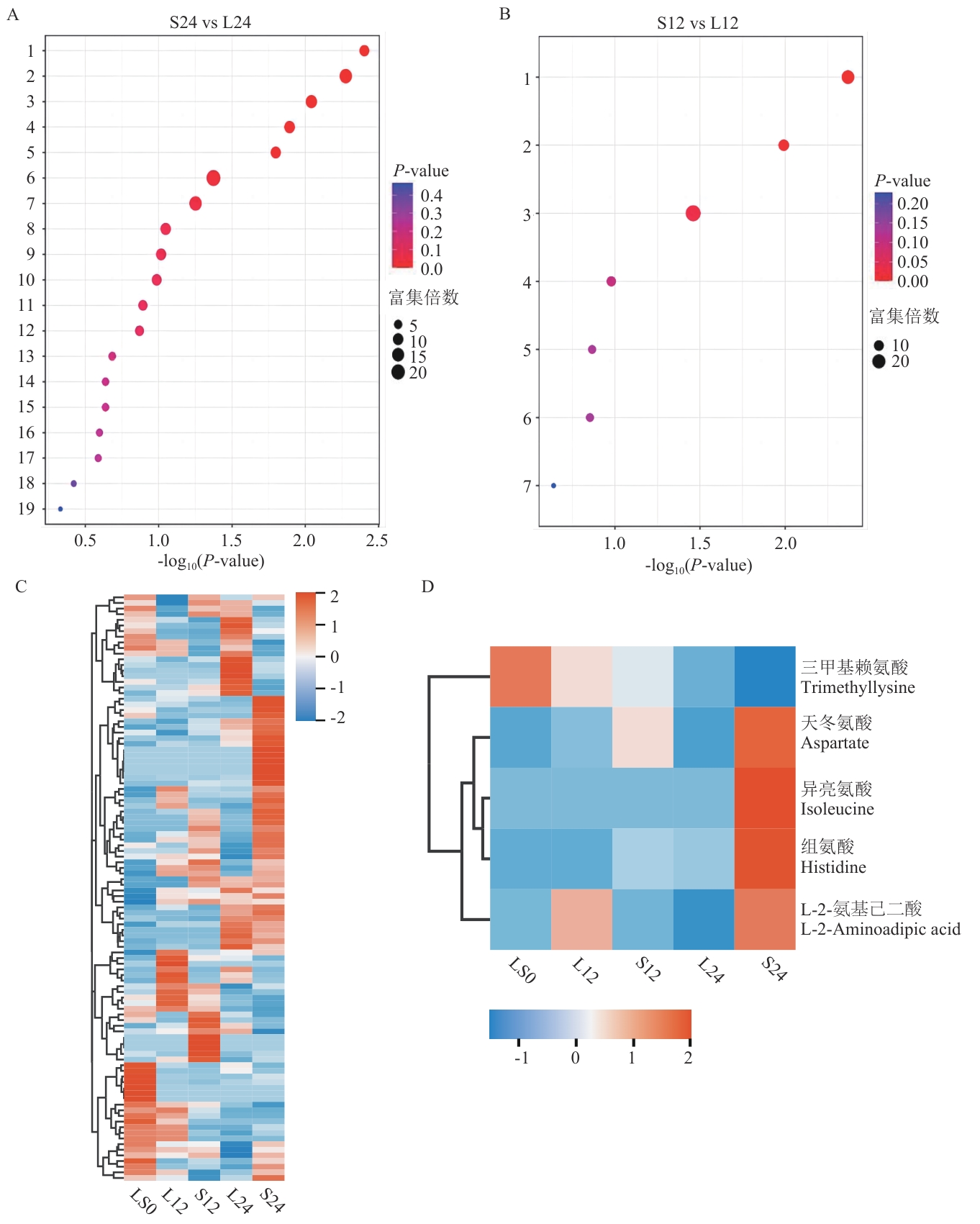
图5 KEGG代谢通路富集气泡图及氨基酸类物质热图A、B:分别为S24 vs L24、L12 vs S12组的KEGG通路富集的气泡图;C:所有氨基酸类代谢物定量聚类热图;D:氨基酸类差异代谢物定量聚类热图。气泡图气泡越大,表示该通路中富集的差异代谢物越多。图A中:1:氨基酸-tRNA生物合成;2:组氨酸代谢;3:β-丙氨酸代谢;4:赖氨酸降解;5:丙氨酸、天冬氨酸和谷氨酸代谢;6:磷酸酯和磷酰化代谢;7:缬氨酸、亮氨酸和异亮氨酸生物合成;8:α-亚麻酸代谢;9:精氨酸生物合成;10:烟酸和烟酰胺代谢;11:泛酸和辅酶A生物合成;12:柠檬酸循环(TCA循环);13:乙二酸和二羧酸代谢;14:甘油磷脂代谢;15:不饱和脂肪酸生物合成;16:缬氨酸、亮氨酸和异亮氨酸降解;17:色氨酸代谢;18:嘌呤代谢;19:类固醇激素生物合成。图B中:1:半乳糖代谢;2:酪氨酸代谢;3:泛醌和其他萜类醌生物合成;4:谷胱甘肽代谢;5:氨基糖和核苷糖代谢;6:精氨酸和脯氨酸代谢;7:嘌呤代谢。图C、D热图均为行标准化及聚类,红色表示上调,蓝色表示下调
Fig. 5 Enrichment bubble plot of KEGG pathways and heatmap of amino acid-related metabolitesA and B: KEGG pathway enrichment bubble plots for S24 vs L24 and L12 vs S12 groups, respectively. C: Quantitative clustering heatmap of all amino acid metabolites. D: Quantitative clustering heatmap of differential amino acid metabolites. In the bubble plots, larger bubbles indicate a higher number of differentially enriched metabolites in that pathway. In Fig. A, 1: aminoacyl-tRNA biosynthesis; 2: histidine metabolism; 3: β-alanine metabolism; 4: lysine degradation; 5: alanine, aspartate, and glutamate metabolism; 6: phosphoester and phosphorylation metabolism; 7: biosynthesis of valine, leucine, and isoleucine; 8: α-linolenic acid metabolism; 9: arginine biosynthesis; 10: nicotinate and nicotinamide metabolism; 11: pantothenate and CoA biosynthesis; 12: citric acid cycle (TCA cycle); 13: oxalate and dicarboxylate metabolism; 14: glycerophospholipid metabolism; 15: unsaturated fatty acid biosynthesis; 16: degradation of valine, leucine, and isoleucine; 17: tryptophan metabolism; 18: purine metabolism; 19: steroid hormone biosynthesis. In Fig. B, 1: galactose metabolism; 2: tyrosine metabolism; 3: biosynthesis of ubiquinone and other terpenoid-quinones; 4: glutathione metabolism; 5: amino sugar and nucleotide sugar metabolism; 6: arginine and proline metabolism; 7: purine metabolism. Fig. C and D: The heatmaps are row-normalized and clustered, with red indicating upregulation and blue indicating downregulation

图6 氨基酸相关KEGG代谢通路图红框代表一条通路,蓝色文字代表在S24中显著上调的差异表达代谢物。虚线表示代谢物间接上下级关系,实线表示直接关系。为所有检测到的代谢物绘制热图,红表示上调,蓝表示下调
Fig. 6 Amino acid-related KEGG metabolic pathway diagramRed boxes indicate pathways, blue shows up-regulated metabolites in S24. Dashed lines indicate indirect superior-subordinate relationships of metabolites, and the solid lines indicate direct relationships. A heat map is drawn for all detected metabolites, red for upregulation, blue for downregulation
| [1] | Osnato M, Cota I, Nebhnani P, et al. Photoperiod control of plant growth: flowering time genes beyond flowering [J]. Front Plant Sci, 2022, 12: 805635. |
| [2] | Mohamed H, Tawfik M, Hussein E, et al. Morphogenic responses of two potato cultivars explants to sucrose, photoperiods and growth regulators [J]. Alfarama J Basic Appl Sci, 2024. |
| [3] | Wafa A, Fekry W, Hassan A, et al. In vitro microtuberization of some potato (Solanum tuberosum L.) cultivars as response to media constituents and photoperiod [J]. Journal of Productivity and Development, 2024, 29(2): 81-98. |
| [4] | Al-Madhagi I. The habit of strawberry flowering is the key for runner propagation, where the photoperiod is the main environmental factor-A review [J]. Adv Hort Sci, 2024, 37(4): 433-449. |
| [5] | Guo L, Plunkert M, Luo X, et al. Developmental regulation of stolon and rhizome [J]. Curr Opin Plant Biol, 2021, 59: 101970. |
| [6] | Lee HG, Jeong YY, Lee H, et al. Arabidopsis HISTONE DEACETYLASE 9 stimulates hypocotyl cell elongation by repressing GIGANTEA expression under short day photoperiod [J]. Front Plant Sci, 2022, 13: 950378. |
| [7] | Xu Y, Koroma AA, Weise SE, et al. Daylength variation affects growth, photosynthesis, leaf metabolism, partitioning, and metabolic fluxes [J]. Plant Physiol, 2023, 194(1): 475-490. |
| [8] | Gendron JM, Staiger D. New horizons in plant photoperiodism [J]. Annu Rev Plant Biol, 2023, 74: 481-509. |
| [9] | Hasterok R, Catalan P, Hazen SP, et al. Brachypodium: 20 years as a grass biology model system; the way forward? [J]. Trends Plant Sci, 2022, 27(10): 1002-1016. |
| [10] | Hasterok R, Marasek A, Donnison IS, et al. Alignment of the genomes of Brachypodium distachyonand temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization [J]. Genetics, 2006, 173(1): 349-362. |
| [11] | Raissig MT, Woods DP. The wild grass Brachypodium distachyon as a developmental model system [J]. Curr Top Dev Biol, 2022, 147: 33-71. |
| [12] | Ludwig E, Sumner J, Berry J, et al. Natural variation in Brachypodium distachyon responses to combined abiotic stresses [J]. Plant J, 2024, 117(6): 1676-1701. |
| [13] | Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction [J]. Physiol Plant, 2006, 126(1): 45-51. |
| [14] | Shulaev V, Cortes D, Miller G, et al. Metabolomics for plant stress response [J]. Physiol Plant, 2008, 132(2): 199-208. |
| [15] | Jänkänpää HJ, Mishra Y, Schröder WP, et al. Metabolic profiling reveals metabolic shifts in Arabidopsis plants grown under different light conditions [J]. Plant Cell Environ, 2012, 35(10): 1824-1836. |
| [16] | Ntagkas N, de Vos RCH, Woltering EJ, et al. Modulation of the tomato fruit metabolome by LED light [J]. Metabolites, 2020, 10(6): 266. |
| [17] | Zhan WM, Guo GH, Cui LH, et al. Combined transcriptome and metabolome analysis reveals the effects of light quality on maize hybrids [J]. BMC Plant Biol, 2023, 23(1): 41. |
| [18] | Djerrab D, Bertrand B, Breitler JC, et al. Photoperiod-dependent transcriptional modifications in key metabolic pathways in Coffea arabica [J]. Tree Physiol, 2021, 41(2): 302-316. |
| [19] | Tusevski O, Petreska Stanoeva J, Stefova M, et al. Phenolic profile of dark-grown and photoperiod-exposed Hypericum perforatum L. hairy root cultures [J]. Sci World J, 2013, 2013: 602752. |
| [20] | Yang CJ, Chen W, Tang DD, et al. Metabolomic and transcriptomic insights into anthocyanin biosynthesis in 'ziyan' tea plants under varied photoperiod and temperature conditions [J]. Agronomy, 2024, 14(1): 56. |
| [21] | Yu JY, Yang Y, Luo LJ, et al. Photoperiod-dependent nutrient accumulation in rice cultivated in plant factories: a comparative metabolomic analysis [J]. Foods, 2024, 13(10): 1544. |
| [22] | Vendruscolo RG, Fagundes MB, Maroneze MM, et al. Scenedesmus obliquus metabolomics: effect of photoperiods and cell growth phases [J]. Bioprocess Biosyst Eng, 2019, 42(5): 727-739. |
| [23] | Hoffman DE, Jonsson P, Bylesjö M, et al. Changes in diurnal patterns within the Populus transcriptome and metabolome in response to photoperiod variation [J]. Plant Cell Environ, 2010, 33(8): 1298-1313. |
| [24] | Kumar A, Singh N, Joshi R. Deciphering the metabolic signatures of Trigonella microgreens as a function of photoperiod and temperature using targeted compound analysis and non-targeted UHPLC-QTOF-IMS based approach [J]. Food Res Int, 2024, 176: 113834. |
| [25] | Atif MJ, Amin B, Ghani MI, et al. Transcriptomic analysis of Allium sativum uncovers putative genes involved in photoperiodic pathway and hormone signaling under long day and short day conditions [J]. Plant Sci, 2021, 313: 111095. |
| [26] | Azevedo RA, Arruda P, Turner WL, et al. The biosynthesis and metabolism of the aspartate derived amino acids in higher plants [J]. Phytochemistry, 1997, 46(3): 395-419. |
| [27] | Artins A, Martins MCM, Meyer C, et al. Sensing and regulation of C and N metabolism-novel features and mechanisms of the TOR and SnRK1 signaling pathways [J]. Plant J, 2024, 118(5): 1268-1280. |
| [28] | Lam HM, Coschigano KT, Oliveira IC, et al. The molecular-genetics of nitrogen assimilation into amino acids in higher plants [J]. Annu Rev Plant Physiol Plant Mol Biol, 1996, 47: 569-593. |
| [29] | Richards NG, Schuster SM. An alternative mechanism for the nitrogen transfer reaction in asparagine synthetase [J]. FEBS Lett, 1992, 313(2): 98-102. |
| [30] | Lam HM, Peng SS, Coruzzi GM. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana [J]. Plant Physiol, 1994, 106(4): 1347-1357. |
| [31] | Lei SH, Rossi S, Huang BR. Metabolic and physiological regulation of aspartic acid-mediated enhancement of heat stress tolerance in perennial ryegrass [J]. Plants, 2022, 11(2): 199. |
| [32] | Han M, Zhang C, Suglo P, et al. L-aspartate: an essential metabolite for plant growth and stress acclimation [J]. Molecules, 2021, 26(7): 1887. |
| [33] | Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants [J]. Amino Acids, 2006, 30(2): 143-162. |
| [34] | Peng C, Uygun S, Shiu SH, et al. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis [J]. Plant Physiol, 2015, 169(3): 1807-1820. |
| [35] | Kochevenko A, Araújo WL, Maloney GS, et al. Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits [J]. Mol Plant, 2012, 5(2): 366-375. |
| [36] | Janeczko A. Estrogens and androgens in plants: the last 20 years of studies [J]. Plants, 2021, 10(12): 2783. |
| [37] | Dembitsky VM. Biological activity and structural diversity of steroids containing aromatic rings, phosphate groups, or halogen atoms [J]. Molecules, 2023, 28(14): 5549. |
| [38] | Khaleel TF, Dillman R, Gretch D. Estradiol distribution during the development and expression of reproductive structures in Populus tremuloides Michx [J]. Sex Plant Reprod, 2003, 16(1): 35-42. |
| [39] | Janeczko A, Filek W, Biesaga-Kościelniak J, et al. The influence of animal sex hormones on the induction of flowering in Arabidopsis thaliana: comparison with the effect of 24-epibrassinolide [J]. Plant Cell Tissue Organ Cult, 2003, 72(2): 147-151. |
| [40] | Janeczko A, Filek W. Stimulation of generative development in partly vernalized winter wheat by animal sex hormones [J]. Acta Physiol Plant, 2002, 24(3): 291-295. |
| [1] | 牛若宇, 高瞻, 熊显鹏, 祝德, 罗皓天, 马学远, 胡冠菁. 棉花野生种质资源的育种应用研究与前景[J]. 生物技术通报, 2025, 41(4): 21-32. |
| [2] | 何财林, 卢晶, 郭会会, 李小安, 吴琪. 藜麦MADS-box基因家族的全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(1): 157-172. |
| [3] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [4] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| [5] | 韩乐乐, 宋文迪, 边嘉珅, 李阳, 杨双胜, 陈紫怡, 李晓薇. 转录组与代谢组联合分析揭示大豆GmERD15c参与盐胁迫下类黄酮的生物合成[J]. 生物技术通报, 2024, 40(10): 243-252. |
| [6] | 姜宇舢, 兰倩, 王芳, 姜亮, 裴成成. 一个影响酪氨酸代谢藜麦突变体的鉴定[J]. 生物技术通报, 2024, 40(10): 253-261. |
| [7] | 牛德, 何跃辉. 季节性因素调控小麦开花时间的分子与表观遗传机制[J]. 生物技术通报, 2024, 40(10): 30-40. |
| [8] | 何诗瑜, 曾仲大, 李博岩. 空间分辨代谢组学在疾病诊断研究中的应用进展[J]. 生物技术通报, 2024, 40(1): 145-159. |
| [9] | 周嫒婷, 彭睿琦, 王芳, 伍建榕, 马焕成. 生防菌株DZY6715在不同生长期的代谢差异分析[J]. 生物技术通报, 2023, 39(9): 225-235. |
| [10] | 韩华蕊, 杨宇琭, 门艺涵, 韩尚玲, 韩渊怀, 霍轶琼, 侯思宇. 基于代谢组学研究谷子SiYABBYs参与花发育过程中鼠李糖苷的生物合成[J]. 生物技术通报, 2023, 39(6): 189-198. |
| [11] | 邢媛, 宋健, 李俊怡, 郑婷婷, 刘思辰, 乔治军. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251. |
| [12] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| [13] | 古丽加马力·艾萨, 邢军, 李安, 张瑞. 开菲尔粒中微生物对苯并(α)芘的非靶向代谢组学分析[J]. 生物技术通报, 2022, 38(5): 123-135. |
| [14] | 朱秋雨, 段绪果. L-天冬氨酸-α-脱羧酶的重组表达、定点突变及高通量检测方法的建立[J]. 生物技术通报, 2022, 38(5): 269-278. |
| [15] | 付雅丽, 彭万里, 林双君, 邓子新, 梁如冰. 香茅醇假单胞菌SJTE-3的短链脱氢酶SDR-X1的克隆及酶性质测定[J]. 生物技术通报, 2022, 38(3): 121-129. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||